Difference between revisions of "Part:BBa K1033003"
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1033003 short</partinfo> | <partinfo>BBa_K1033003 short</partinfo> | ||
| − | + | An assembly of our two biobricks, 4Cl and STS. This translational fusion can produce resveratrol from p-coumaric acid. [1] | |
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/d/d1/Uppsala_4CL-STS.png | ||
| + | |||
| + | '''Application''' | ||
| + | |||
| + | This biobrick can be used to produce the healthy antioxidant resveratrol form p-coumaric acid. Togehter with our biobrick TAL: [https://parts.igem.org/Part:BBa_K1033000 BBa_K1033000], we can produce resveratrol directly from tyrosine. [1] | ||
| + | |||
| + | '''Characterizaion''' | ||
| + | |||
| + | This biobrick can be characterized with for example hplc. | ||
| + | |||
| + | |||
| + | == Characterizaion data == | ||
| + | |||
| + | '''High pressure liquid chromatography''' | ||
| + | |||
| + | We tested our biobrick 4Cl-STS on HPLC, by adding p-coumaric acid as a precursor. The result we saw was quite unclear. We saw that the E. coli produced something out of the ordinary, but the absorbation was low and the peaks did not exactly match the standard. The peak at around ~33 min could correspond to our standard, but it is unclear. We theorize it is something in the actual hplc measurement that fails, or that something happens to our resveratrol metabolite in our e-coli. This result could correspond to the poor results in our blot. We hope that iGEM teams can continue to work on these biobricks in the future. | ||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/d/dc/Resva_uppsala_tab1.png | ||
| + | |||
| + | ''Figure 2: E. coli supposed to produce resveratrol. As we can see, we got very low absorbance peaks at ~30 min, ~33 min and ~36 min.'' | ||
| + | |||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/3/33/Resva_uppsala_tab2.png | ||
| + | |||
| + | ''Figure 3: Resveratrol standard, peaks around ~33, ~34 min.'' | ||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/8/82/Resva_uppsala_tab3.png | ||
| + | |||
| + | ''Figure 4: Resveratrol standard that is scaled down to correspond to the absorbations of our e. coli supposed to produce the corresponding metabolite. The peaks are at around ~33 and ~34.'' | ||
| + | |||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/4/4f/Uppsala_char_coumaric-acid_blank.png | ||
| + | |||
| + | ''Figure 5: E. coli culture injected to the hplc without our biobrick tyrosine ammonia lyase. Here we can see that there is originally no peaks around 30-35 minutes. | ||
| + | '' | ||
| + | |||
| + | '''References''' | ||
| + | |||
| + | |||
| + | [1] Robert J. Conrado et al, DNA guided assembly of biosynthetic pathways promotes improved catalytic effiency. Nucleic Acids Research , 2012, Vol 40 NO 4, 1879-1889 | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 23:48, 4 October 2013
B0034-4Cl-B0034-STS
An assembly of our two biobricks, 4Cl and STS. This translational fusion can produce resveratrol from p-coumaric acid. [1]
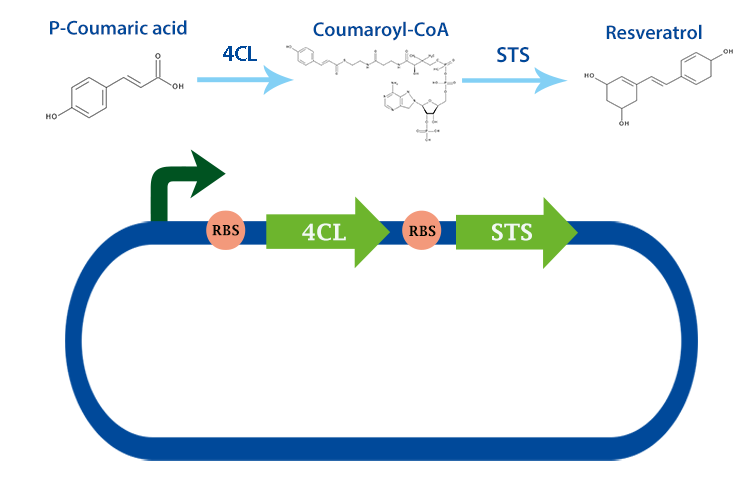
Application
This biobrick can be used to produce the healthy antioxidant resveratrol form p-coumaric acid. Togehter with our biobrick TAL: BBa_K1033000, we can produce resveratrol directly from tyrosine. [1]
Characterizaion
This biobrick can be characterized with for example hplc.
Characterizaion data
High pressure liquid chromatography
We tested our biobrick 4Cl-STS on HPLC, by adding p-coumaric acid as a precursor. The result we saw was quite unclear. We saw that the E. coli produced something out of the ordinary, but the absorbation was low and the peaks did not exactly match the standard. The peak at around ~33 min could correspond to our standard, but it is unclear. We theorize it is something in the actual hplc measurement that fails, or that something happens to our resveratrol metabolite in our e-coli. This result could correspond to the poor results in our blot. We hope that iGEM teams can continue to work on these biobricks in the future.

Figure 2: E. coli supposed to produce resveratrol. As we can see, we got very low absorbance peaks at ~30 min, ~33 min and ~36 min.

Figure 3: Resveratrol standard, peaks around ~33, ~34 min.
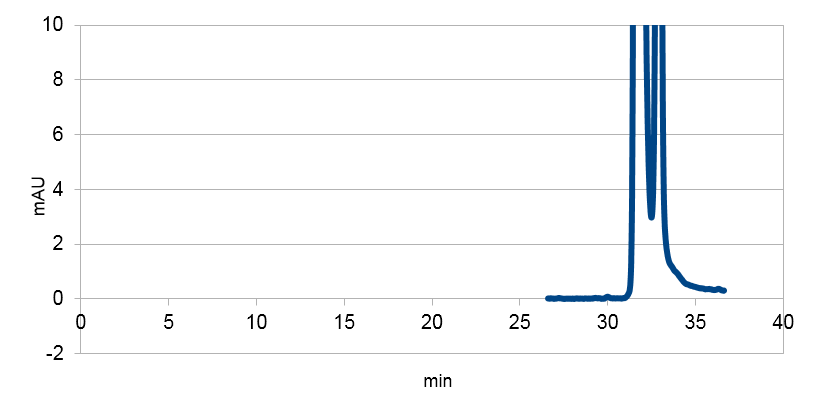
Figure 4: Resveratrol standard that is scaled down to correspond to the absorbations of our e. coli supposed to produce the corresponding metabolite. The peaks are at around ~33 and ~34.
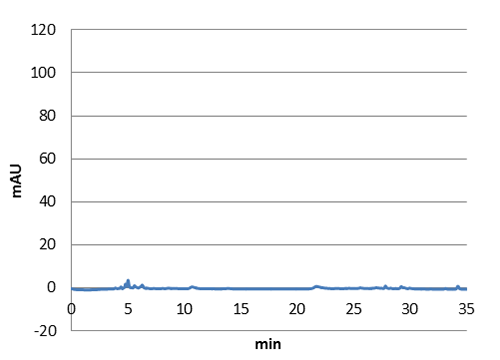
Figure 5: E. coli culture injected to the hplc without our biobrick tyrosine ammonia lyase. Here we can see that there is originally no peaks around 30-35 minutes.
References
[1] Robert J. Conrado et al, DNA guided assembly of biosynthetic pathways promotes improved catalytic effiency. Nucleic Acids Research , 2012, Vol 40 NO 4, 1879-1889
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1108
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1675
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1307
