Difference between revisions of "Part:BBa K1033002"
| Line 10: | Line 10: | ||
| + | == Characterizaion data == | ||
| + | |||
| + | |||
| + | '''Summary''' | ||
| + | |||
| + | Although we managed to clone out and sequence verify the genes for resveratrol production, we have had some problems in the characterization. The results are unclear, and we did not have time for further investigations.For detailed information about the characterisation methods, see the protocol section | ||
| + | |||
| + | '''Biobrick''' | ||
| + | |||
| + | We managed to biobrick and sequence verify this part. | ||
| + | |||
| + | '''Western blot''' | ||
| + | |||
| + | We also succeeded in expressing the enzyme stilbene synthase in E. coli. Although our expression of the protein was very weak, and due to time constraints we were not able to optimize our experiment. | ||
| + | |||
| + | To enable the detection of this protein by anti-his antibodies, 6-histidine tags was incorporated in the sequence. This way we could detect our enzyme with anti-his antibodies. | ||
| + | |||
| + | We expressed our protein with a promotor working in both lactobacillus and e-coli. This way, we can easily transfer stilbene synthase to lactobacillus later on. | ||
| + | |||
| + | The size of our protein was calculated using ProtParam [5], 43 kDA. | ||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/3/39/Resv_westblot_uppsala.png | ||
| + | |||
| + | ''Figure 1:Number 2 shows a very weak band of our protein at around 43 kDA. | ||
| + | Positive control -> 1, Stilbene synthase -> 2'' | ||
| + | |||
| + | |||
| + | |||
| + | '''High pressure liquid chromatography''' | ||
| + | |||
| + | We tested our biobrick 4Cl-STS on HPLC, by adding p-coumaric acid as a precursor. The result we saw was quite unclear. We saw that the e-coli produced something out of the ordinary, but the absorbation was low and the peaks did not exactly match the standard. The peak at around ~33 min could correspond to our standard, but it is unclear. We theorize it is something in the actual hplc measurement that fails, or that something happens to our resveratrol metabolite in our e-coli. This result could correspond to the poor results in our blot. We hope that iGEM teams can continue to work on these biobricks in the future. | ||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/d/dc/Resva_uppsala_tab1.png | ||
| + | |||
| + | ''Figure 2: E. coli supposed to produce resveratrol. As we can see, we got very low absorbance peaks at ~30 min, ~33 min and ~36 min.'' | ||
| + | |||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/3/33/Resva_uppsala_tab2.png | ||
| + | |||
| + | ''Figure 3: Resveratrol standard, peaks around ~33, ~34 min.'' | ||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/8/82/Resva_uppsala_tab3.png | ||
| + | |||
| + | ''Figure 4: Resveratrol standard that is scaled down to correspond to the absorbations of our e. coli supposed to produce the corresponding metabolite. The peaks are at around ~33 and ~34.'' | ||
| + | |||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2013/4/4f/Uppsala_char_coumaric-acid_blank.png | ||
| + | |||
| + | ''Figure 5: E. coli culture injected to the hplc without our biobrick tyrosine ammonia lyase. Here we can see that there is originally no peaks around 30-35 minutes. | ||
| + | '' | ||
| + | |||
| + | '''References''' | ||
| + | |||
| + | [1] Sinclair, D. A & Baur, Y, A. Therapeutic potential of resveratrol: the in vivo evidence. Nature 506 | JUNE 2006 | VOLUME 5 | ||
| + | |||
| + | [2] Baur, Y, A. et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature Vol 444| 16 November 2006 | ||
| + | |||
| + | [3] Robert J. Conrado et al, DNA guided assembly of biosynthetic pathways promotes improved catalytic effiency. Nucleic Acids Research , 2012, Vol 40 NO 4, 1879-1889 | ||
| + | |||
| + | [4] Sinclair, D. A & Baur, Y, A. Therapeutic potential of resveratrol: the in vivo evidence. Nature 506 | JUNE 2006 | VOLUME 5 | ||
| + | |||
| + | [5] Baur, Y, A. et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature Vol 444| 16 November 2006 | ||
| + | |||
| + | [6] Here's a link to ProtParam: http://web.expasy.org/protparam/ | ||
Revision as of 17:17, 4 October 2013
stilbene synthase (STS) with RBS
This part codes for the enzyme stilbene synthase (STS). Stilbene synthase is the third and last enzyme in the stilbene synthesis pathway which derrives from the universal polyphenol biosynthetic pathway. It catalyzes the formation of resveratrol, using the substrates coumaryl-CoA and malonyl-CoA.[1]
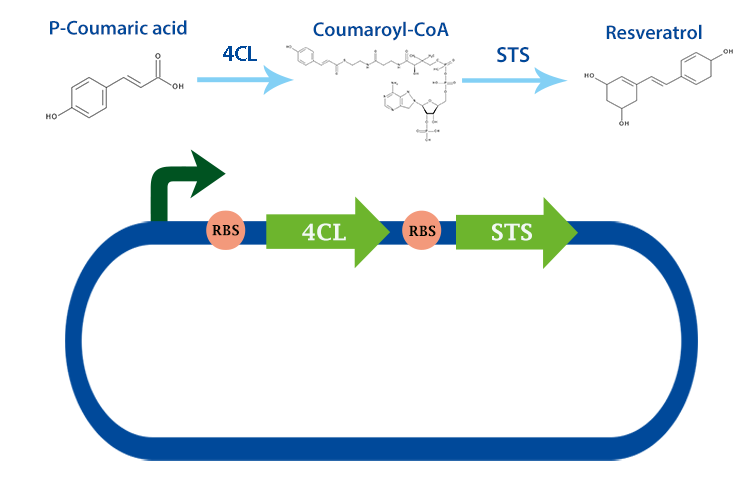
Applications With the help of this enzyme one can produce the healthy antioxidant resveratrol derived from redwhine. For the production of resveratrol, coumaryl-CoA can be obtained from our other biobrick, 4Cl.
Characterizaion data
Summary
Although we managed to clone out and sequence verify the genes for resveratrol production, we have had some problems in the characterization. The results are unclear, and we did not have time for further investigations.For detailed information about the characterisation methods, see the protocol section
Biobrick
We managed to biobrick and sequence verify this part.
Western blot
We also succeeded in expressing the enzyme stilbene synthase in E. coli. Although our expression of the protein was very weak, and due to time constraints we were not able to optimize our experiment.
To enable the detection of this protein by anti-his antibodies, 6-histidine tags was incorporated in the sequence. This way we could detect our enzyme with anti-his antibodies.
We expressed our protein with a promotor working in both lactobacillus and e-coli. This way, we can easily transfer stilbene synthase to lactobacillus later on.
The size of our protein was calculated using ProtParam [5], 43 kDA.

Figure 1:Number 2 shows a very weak band of our protein at around 43 kDA. Positive control -> 1, Stilbene synthase -> 2
High pressure liquid chromatography
We tested our biobrick 4Cl-STS on HPLC, by adding p-coumaric acid as a precursor. The result we saw was quite unclear. We saw that the e-coli produced something out of the ordinary, but the absorbation was low and the peaks did not exactly match the standard. The peak at around ~33 min could correspond to our standard, but it is unclear. We theorize it is something in the actual hplc measurement that fails, or that something happens to our resveratrol metabolite in our e-coli. This result could correspond to the poor results in our blot. We hope that iGEM teams can continue to work on these biobricks in the future.

Figure 2: E. coli supposed to produce resveratrol. As we can see, we got very low absorbance peaks at ~30 min, ~33 min and ~36 min.

Figure 3: Resveratrol standard, peaks around ~33, ~34 min.
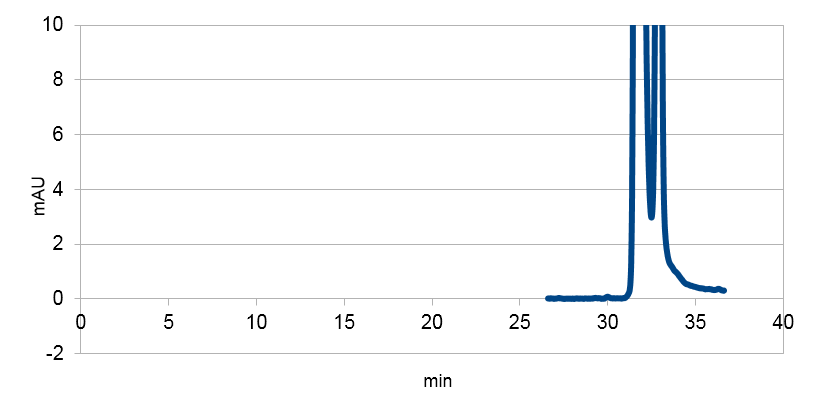
Figure 4: Resveratrol standard that is scaled down to correspond to the absorbations of our e. coli supposed to produce the corresponding metabolite. The peaks are at around ~33 and ~34.
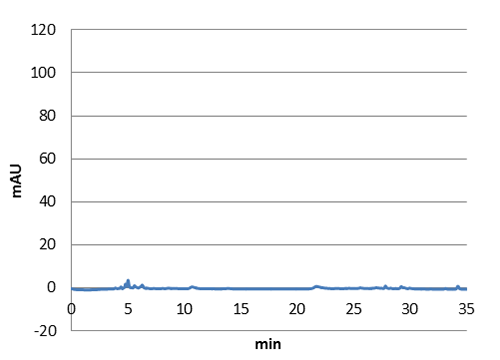
Figure 5: E. coli culture injected to the hplc without our biobrick tyrosine ammonia lyase. Here we can see that there is originally no peaks around 30-35 minutes.
References
[1] Sinclair, D. A & Baur, Y, A. Therapeutic potential of resveratrol: the in vivo evidence. Nature 506 | JUNE 2006 | VOLUME 5
[2] Baur, Y, A. et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature Vol 444| 16 November 2006
[3] Robert J. Conrado et al, DNA guided assembly of biosynthetic pathways promotes improved catalytic effiency. Nucleic Acids Research , 2012, Vol 40 NO 4, 1879-1889
[4] Sinclair, D. A & Baur, Y, A. Therapeutic potential of resveratrol: the in vivo evidence. Nature 506 | JUNE 2006 | VOLUME 5
[5] Baur, Y, A. et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature Vol 444| 16 November 2006
[6] Here's a link to ProtParam: http://web.expasy.org/protparam/
1. Chin Giaw Lim, Zachary L. Fowler, Thomas Hueller, Steffen Schaffer, Mattheos A. G. Koffas (2011) High-Yield Resveratrol Production in Engineered Escherichia coli. Appl Environ Microbiol 77(10):3451
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
