Difference between revisions of "Part:BBa I13453:Experience"
(→User Reviews) |
(→User Reviews) |
||
| (6 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | ||
| Line 18: | Line 17: | ||
|}; | |}; | ||
<!-- End of the user review template --> | <!-- End of the user review template --> | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_I13453 AddReview 5</partinfo> | ||
| + | <I>Patrick Rosendahl Andreassen</I> | ||
| + | |width='60%' valign='top'| | ||
| + | To test if overexpression of AraC improved expression control of the arabinose promoter on high-copy plasmids, devices with and without a araC device (BBa_K1088017) was assayed. Duplicates of MG1655 strains carrying either pSB1C3-Para-HRT2 (BBa_K1088024; -araC) or pSB1C3-Pcon-araC-term-Para-HRT2 (BBa_K1088016; +araC) were grown to late-exponential phase: OD<sub>600</sub>=0.8. At this OD the strains were induced with 0.2 % arabinose at time t=0 min, and samples were taken at times: -2 min, 15 min, and 30 min. Total RNA purified from the samples were run on a gel, blotted onto a membrane, and hybridized with probes specific for ''HRT2'' mRNA and 5S rRNA (loading control), respectively. | ||
| + | |||
| + | https://static.igem.org/mediawiki/2013/e/ee/SDU2013_Part_BBa_K1088017.png | ||
| + | |||
| + | A) Normalized intensity of ''HRT2'' mRNA using intensity of 5S rRNA as reference. The normalized intensity of sample -araC -2min were set to 1 and the other samples are relative to that. Within 15 min of induction the expression levels are at its maximum in both strains, and overexpression of AraC does not seem to be necessary for expression control of the arabinose promoter. Though, we cannot conclude that it has no effect in the minutes prior to 15 min after induction | ||
| + | B) Northern blot result reflecting diagram. | ||
| + | |||
| + | See detailed protocol at http://2013.igem.org/Team:SDU-Denmark/Tour41 protocol #55 | ||
| + | |} | ||
| + | |||
{|width='80%' style='border:1px solid gray' | {|width='80%' style='border:1px solid gray' | ||
|- | |- | ||
| Line 33: | Line 48: | ||
<I>kahaynes</I> | <I>kahaynes</I> | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| − | '''pBad I13453''' confers expression of tetracycline resistance when placed upstream of '''hixC-RBS-Tet...''' (<partinfo>BBa_J44010</partinfo>). The presence of '''hixC''' (26 bp) between '''pBad''' and '''RBS-Tet''' does not disrupt transcription. In the insulated vector <partinfo>pSB1A7</partinfo> we have observed that tetracycline resistance depends upon the presence and proper orientation of '''pBad''' relative to the coding region (Davidson and Missouri Western 2006). '''pBad-hixC-RBS-Tet''' is tetracycline resistant, whereas '''RBS-Tet''' and '''pBad-hixC-Tet<sub>rev</sub>-RBS<sub>rev</sub>''' are not. We have observed Tet expression in the absence of L-arabinose (the affects of L-arabinose were not tested in this system). In summary, '''pBad I13453''' (which includes the Cap and AraC protein binding domains and polymerase binding site; Lee, PNAS, 1981) appears to be a functional constitutive promoter. | + | '''pBad I13453''' confers expression of tetracycline resistance when placed upstream of '''hixC-RBS-Tet...''' (<partinfo>BBa_J44010</partinfo>). The presence of '''hixC''' (26 bp) between '''pBad''' and '''RBS-Tet''' does not disrupt transcription. In the insulated vector <partinfo>pSB1A7</partinfo> we have observed that tetracycline resistance depends upon the presence and proper orientation of '''pBad''' relative to the coding region (Davidson and Missouri Western 2006). '''pBad-hixC-RBS-Tet''' is tetracycline resistant, whereas '''RBS-Tet''' and '''pBad-hixC-Tet<sub>rev</sub>-RBS<sub>rev</sub>''' are not. We have observed Tet expression in the absence of L-arabinose (the affects of L-arabinose were not tested in this system). In summary, '''pBad I13453''' (which includes the Cap and AraC protein binding domains and polymerase binding site; Lee, PNAS, 1981) appears to be a functional constitutive promoter that does not depend upon L-arabinose for expression. |
|} | |} | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_I13453 AddReview 5</partinfo> | ||
| + | <I>British_Columbia iGEM 2009</I> | ||
| + | |width='60%' valign='top'| | ||
| + | We performed extensive testing on <partinfo>I13453</partinfo> as part of our creation of a family of variable-strength arabinose-inducible promoters. We found that induction of <partinfo>I13453</partinfo> with L-arabinose led to expression of a coupled fluorescent reporter; contrary to the above review, we did not observe any reporter expression in the absence of L-arabinose. <partinfo>I13453</partinfo> response to L-arabinose was specific and dependent on input concentration. Our detailed experimental measurements for <partinfo>I13453</partinfo> can be found [[Part:BBa_I13453:British_Columbia|here]] and a comparison of the entire pBAD promoter family can be found [https://parts.igem.org/PBAD_Promoter_Family here]. | ||
| + | |} | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K584001 AddReview 5</partinfo> | ||
| + | <I><B>[http://2011.igem.org/Team:KULeuven K.U.Leuven iGEM 2011 Team]</B></I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | <u><b>Characterization by K.U.Leuven 2011 iGEM Team</b></u> | ||
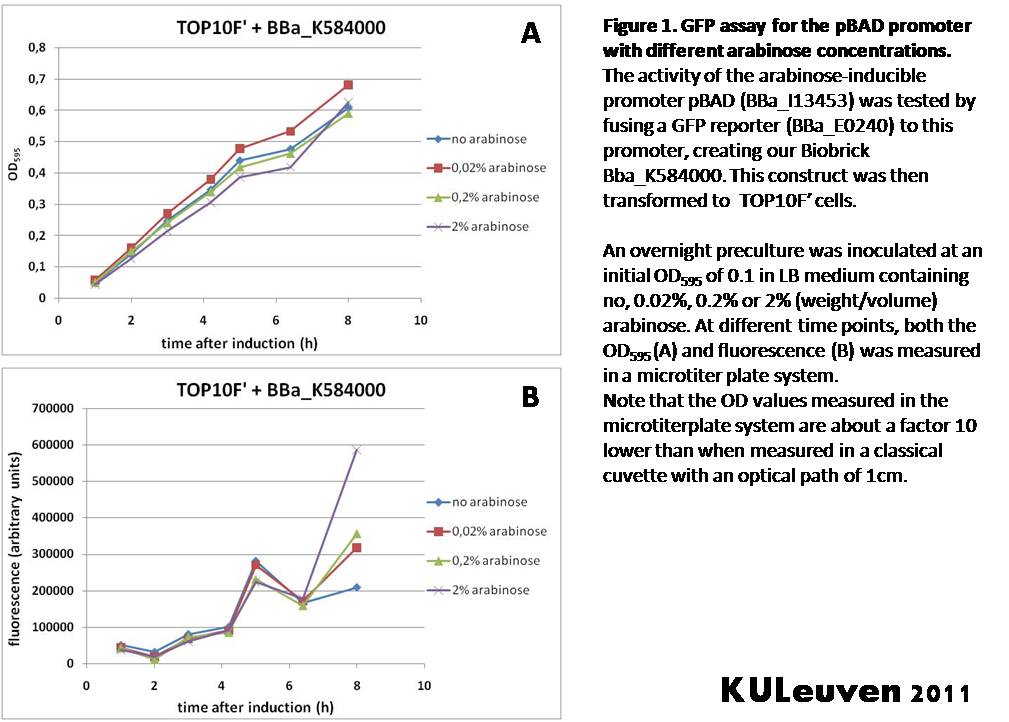
| + | <p align="justify">To test the usefulness of the arabinose-inducible promoter I13453 in our 2011 iGEM project, we fused the promoter to a GFP reporter, and assayed the promoter’s activity after addition of different amounts of arabinose. We tested this activity both in a TOP10F’ (Figure 1) as well as a MG1655 (Figure 2) E.coli strain background. For more information on E.coli strain descriptions, we recommend the following web site: [http://openwetware.org/wiki/E._coli_genotypes E.Coli_Genotypes].</p> | ||
| + | |||
| + | [[Image:pBAD_TOP10.jpg|thumb|center|Figure1]] | ||
| + | |||
| + | |||
| + | <p align="justify">As can be seen in Figure 1, addition of arabinose had only minor, if any, effect on the growth of TOP10F’ cells, most likely because these cells cannot metabolize the sugar. However, to our surprise, we did not observe an arabinose-induced increase of fluorescence compared to the control without arabinose for the first 6 hours. The observed increase in fluorescence after arabinose addition at the 8 hour time point could not be confirmed when the experiment was repeated.</p> | ||
| + | |||
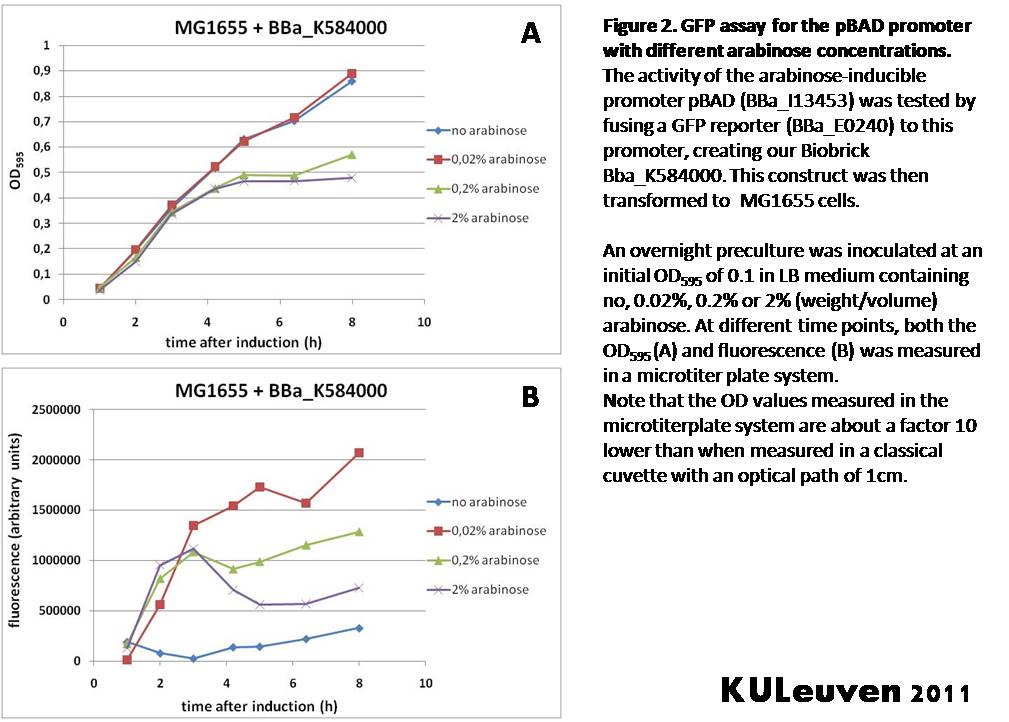
| + | [[Image:pBAD_MG1655.jpg|thumb|center|Figure2]] | ||
| + | |||
| + | |||
| + | <p align="justify">Figure 2 demonstrates that in MG1655 cells, the addition of 0.2% and 2% arabinose results in a growth defect after about 4 hours of growth, which may relate to the metabolism of the sugar in this strain. In contrast to the TOP10F’ cells, we see a clear induction of promoter activity after addition of arabinose. The addition of only 0.02% resulted in the greatest induction. Increasing arabinose concentration did not increase fluorescence, probably due to the observed growth defect. </p> | ||
| + | |||
| + | |||
| + | <p align="justify">As an additional control, we checked the activity of a constitutive promoter under the same conditions as described here. For results on these experiments, check out our [https://parts.igem.org/wiki/index.php?title=Part:BBa_K584001 BBa_K584001] page.</p> | ||
| + | |||
| + | |} | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_I13453 AddReview 1</partinfo> | ||
| + | <I>Michigan iGEM 2012</I> | ||
| + | |width='60%' valign='top'| | ||
| + | According to sequence data, <partinfo>BBa_I13453</partinfo> is missing O2 site necessary for repression by AraC. (Schleif R. AraC protein: a love-hate relationship. Bioessays 2003;25:274-282.) | ||
| + | |} | ||
| + | |||
| + | |||
<!-- DON'T DELETE --><partinfo>BBa_I13453 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_I13453 EndReviews</partinfo> | ||
Latest revision as of 19:05, 3 October 2013
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_I13453
User Reviews
UNIQc6c5a210b511e078-partinfo-00000000-QINU
|
•••••
Patrick Rosendahl Andreassen |
To test if overexpression of AraC improved expression control of the arabinose promoter on high-copy plasmids, devices with and without a araC device (BBa_K1088017) was assayed. Duplicates of MG1655 strains carrying either pSB1C3-Para-HRT2 (BBa_K1088024; -araC) or pSB1C3-Pcon-araC-term-Para-HRT2 (BBa_K1088016; +araC) were grown to late-exponential phase: OD600=0.8. At this OD the strains were induced with 0.2 % arabinose at time t=0 min, and samples were taken at times: -2 min, 15 min, and 30 min. Total RNA purified from the samples were run on a gel, blotted onto a membrane, and hybridized with probes specific for HRT2 mRNA and 5S rRNA (loading control), respectively.
A) Normalized intensity of HRT2 mRNA using intensity of 5S rRNA as reference. The normalized intensity of sample -araC -2min were set to 1 and the other samples are relative to that. Within 15 min of induction the expression levels are at its maximum in both strains, and overexpression of AraC does not seem to be necessary for expression control of the arabinose promoter. Though, we cannot conclude that it has no effect in the minutes prior to 15 min after induction B) Northern blot result reflecting diagram. See detailed protocol at http://2013.igem.org/Team:SDU-Denmark/Tour41 protocol #55 |
|
•••••
Antiquity |
This review comes from the old result system and indicates that this part worked in some test. |
|
•••••
kahaynes |
pBad I13453 confers expression of tetracycline resistance when placed upstream of hixC-RBS-Tet... (BBa_J44010). The presence of hixC (26 bp) between pBad and RBS-Tet does not disrupt transcription. In the insulated vector pSB1A7 we have observed that tetracycline resistance depends upon the presence and proper orientation of pBad relative to the coding region (Davidson and Missouri Western 2006). pBad-hixC-RBS-Tet is tetracycline resistant, whereas RBS-Tet and pBad-hixC-Tetrev-RBSrev are not. We have observed Tet expression in the absence of L-arabinose (the affects of L-arabinose were not tested in this system). In summary, pBad I13453 (which includes the Cap and AraC protein binding domains and polymerase binding site; Lee, PNAS, 1981) appears to be a functional constitutive promoter that does not depend upon L-arabinose for expression. |
|
•••••
British_Columbia iGEM 2009 |
We performed extensive testing on BBa_I13453 as part of our creation of a family of variable-strength arabinose-inducible promoters. We found that induction of BBa_I13453 with L-arabinose led to expression of a coupled fluorescent reporter; contrary to the above review, we did not observe any reporter expression in the absence of L-arabinose. BBa_I13453 response to L-arabinose was specific and dependent on input concentration. Our detailed experimental measurements for BBa_I13453 can be found here and a comparison of the entire pBAD promoter family can be found here. |
|
•••••
[http://2011.igem.org/Team:KULeuven K.U.Leuven iGEM 2011 Team] |
Characterization by K.U.Leuven 2011 iGEM Team To test the usefulness of the arabinose-inducible promoter I13453 in our 2011 iGEM project, we fused the promoter to a GFP reporter, and assayed the promoter’s activity after addition of different amounts of arabinose. We tested this activity both in a TOP10F’ (Figure 1) as well as a MG1655 (Figure 2) E.coli strain background. For more information on E.coli strain descriptions, we recommend the following web site: [http://openwetware.org/wiki/E._coli_genotypes E.Coli_Genotypes].
As can be seen in Figure 1, addition of arabinose had only minor, if any, effect on the growth of TOP10F’ cells, most likely because these cells cannot metabolize the sugar. However, to our surprise, we did not observe an arabinose-induced increase of fluorescence compared to the control without arabinose for the first 6 hours. The observed increase in fluorescence after arabinose addition at the 8 hour time point could not be confirmed when the experiment was repeated.
Figure 2 demonstrates that in MG1655 cells, the addition of 0.2% and 2% arabinose results in a growth defect after about 4 hours of growth, which may relate to the metabolism of the sugar in this strain. In contrast to the TOP10F’ cells, we see a clear induction of promoter activity after addition of arabinose. The addition of only 0.02% resulted in the greatest induction. Increasing arabinose concentration did not increase fluorescence, probably due to the observed growth defect.
As an additional control, we checked the activity of a constitutive promoter under the same conditions as described here. For results on these experiments, check out our BBa_K584001 page. |
|
•
Michigan iGEM 2012 |
According to sequence data, BBa_I13453 is missing O2 site necessary for repression by AraC. (Schleif R. AraC protein: a love-hate relationship. Bioessays 2003;25:274-282.) |
UNIQc6c5a210b511e078-partinfo-0000000E-QINU