Difference between revisions of "Part:BBa K1051206"
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1051206 short</partinfo> | <partinfo>BBa_K1051206 short</partinfo> | ||
| − | + | <h3>Purpose</h3> | |
| − | + | SsrA degradation tag in E. <i>coli</i>, add TAATAA to C-terminal of Biobrick M0050. | |
| − | <!-- Add more about the biology of this part | + | <br /> |
| + | <h3>Principle</h3> | ||
| + | '''SsrA degradation tag'''. In E. <i>coli</i>, the adaptor SspB tethers ssrAtagged substrates to the ClpXP protease, causing a modest increase in their rate of degradation. Which means, a variation of the WT SsrA tag sequence will accelerate the degradation of proteins when fused to their C-terminal. Thus the degradation rates are dependent on concentration of proteases and binding mediators. In order to fuse degradation tags freely on the C-terminal of protein, we add TAATAA to the tail of M0050 to construnt this part(see detailed about M0050 at https://parts.igem.org/Part:BBa_M0050).<br /> | ||
| + | We constructed the measurement pathway of tag K1051257(contains J04500, K1051000 and K1051206) to test the rates of degradation of tagged proteins. J04450 was used as positive control because of the same promoter and fluorescent protein. | ||
| + | <br /> | ||
| + | <!-- Add more about the biology of this part her | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
<!-- --> | <!-- --> | ||
| − | < | + | |
| + | <h3>Measurement</h3> | ||
| + | https://static.igem.org/mediawiki/2013/6/68/Growth-Curve.jpg | ||
| + | The growth curve of E. <i>coli</i>. | ||
| + | https://static.igem.org/mediawiki/2013/e/e9/Degladder.jpg | ||
| + | From right to left, the negative control,BBa_K1051257, BBa_K1051258, BBa_K1051259, J04450 as Positive Control. As the pictures showed, the lights of RFP within three degradation tags are decreasing. | ||
| + | https://static.igem.org/mediawiki/2013/thumb/4/48/Flo3.jpg/800px-Flo3.jpg | ||
| + | The test results of BBa_K1051258. A:No exciting lights; B. Powerful exciting lights; C. Weak exciting lights. | ||
| + | In picture, there are only obvious lights in the picture B, indicated the degradation rates are working | ||
| + | https://static.igem.org/mediawiki/2013/a/a8/Degmicro.jpg | ||
| + | The test results of BBa_K1051258 in chip. A,LB medium,O minuts; B, IPTG medium,9minutes; C,IPTG medium, 15 minutes | ||
| + | <br /> | ||
| + | <br /> | ||
| + | https://static.igem.org/mediawiki/parts/1/1e/Average_flurescence_intensity_of_K1051258_measurement.jpg | ||
| + | The average fluorescence intensity of K1051258 when added IPTG after specific time. | ||
| + | https://static.igem.org/mediawiki/2013/4/47/Degradation_rate_and_fluorescence_intensity.jpg | ||
| + | <br /> | ||
| + | The fluorescence intensity and degradation rate of <i>E</i>. <i>coli</i>. | ||
| + | https://static.igem.org/mediawiki/2013/thumb/a/ad/DT-half_life_and_DR.png/800px-DT-half_life_and_DR.png | ||
| + | <br /> | ||
| + | The data we calculated from microscope and microfluidic mate very well. | ||
| + | <h3>Sequence and Features</h3> | ||
<partinfo>BBa_K1051206 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1051206 SequenceAndFeatures</partinfo> | ||
| − | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
| − | <partinfo> | + | <partinfo>BBa_K1051500 parameters</partinfo> |
<!-- --> | <!-- --> | ||
| + | |||
| + | <h3>References</h3> | ||
| + | [1]McGinness, Baker, Sauer. 2006. Mol. Cell. 22:701.<br /> | ||
| + | [2]Flynn et al 2003. Mol. Cell. 11: 671. Flynn et al. 2001. PNAS 98(19): 10584. Anderson et al 1998. App. Env. Microbiol. 64(6):2240 | ||
Latest revision as of 08:18, 3 October 2013
The degradation tag in E. coil ,M0050 with TAATAA.
Purpose
SsrA degradation tag in E. coli, add TAATAA to C-terminal of Biobrick M0050.
Principle
SsrA degradation tag. In E. coli, the adaptor SspB tethers ssrAtagged substrates to the ClpXP protease, causing a modest increase in their rate of degradation. Which means, a variation of the WT SsrA tag sequence will accelerate the degradation of proteins when fused to their C-terminal. Thus the degradation rates are dependent on concentration of proteases and binding mediators. In order to fuse degradation tags freely on the C-terminal of protein, we add TAATAA to the tail of M0050 to construnt this part(see detailed about M0050 at https://parts.igem.org/Part:BBa_M0050).
We constructed the measurement pathway of tag K1051257(contains J04500, K1051000 and K1051206) to test the rates of degradation of tagged proteins. J04450 was used as positive control because of the same promoter and fluorescent protein.
Measurement
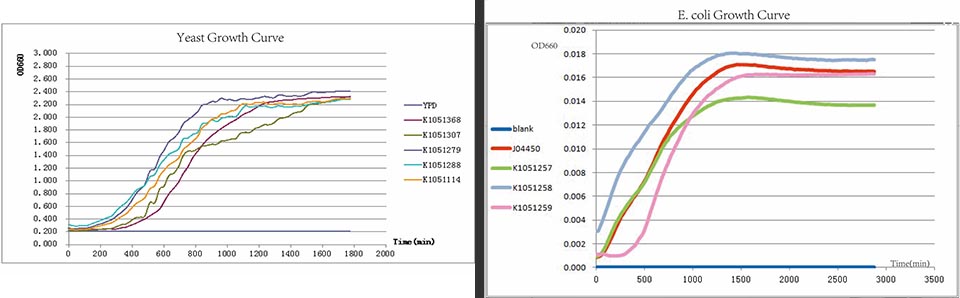 The growth curve of E. coli.
The growth curve of E. coli.
 From right to left, the negative control,BBa_K1051257, BBa_K1051258, BBa_K1051259, J04450 as Positive Control. As the pictures showed, the lights of RFP within three degradation tags are decreasing.
From right to left, the negative control,BBa_K1051257, BBa_K1051258, BBa_K1051259, J04450 as Positive Control. As the pictures showed, the lights of RFP within three degradation tags are decreasing.
 The test results of BBa_K1051258. A:No exciting lights; B. Powerful exciting lights; C. Weak exciting lights.
In picture, there are only obvious lights in the picture B, indicated the degradation rates are working
The test results of BBa_K1051258. A:No exciting lights; B. Powerful exciting lights; C. Weak exciting lights.
In picture, there are only obvious lights in the picture B, indicated the degradation rates are working
 The test results of BBa_K1051258 in chip. A,LB medium,O minuts; B, IPTG medium,9minutes; C,IPTG medium, 15 minutes
The test results of BBa_K1051258 in chip. A,LB medium,O minuts; B, IPTG medium,9minutes; C,IPTG medium, 15 minutes
 The average fluorescence intensity of K1051258 when added IPTG after specific time.
The average fluorescence intensity of K1051258 when added IPTG after specific time.

The fluorescence intensity and degradation rate of E. coli.

The data we calculated from microscope and microfluidic mate very well.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
[1]McGinness, Baker, Sauer. 2006. Mol. Cell. 22:701.
[2]Flynn et al 2003. Mol. Cell. 11: 671. Flynn et al. 2001. PNAS 98(19): 10584. Anderson et al 1998. App. Env. Microbiol. 64(6):2240
