Difference between revisions of "Part:BBa K1041002"
| Line 6: | Line 6: | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K1041002 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | <!-- Uncomment this to enable Functional Parameter display | ||
| + | ===Functional Parameters=== | ||
| + | <partinfo>BBa_K1041002 parameters</partinfo> | ||
| + | <!-- --> | ||
===Characterisation=== | ===Characterisation=== | ||
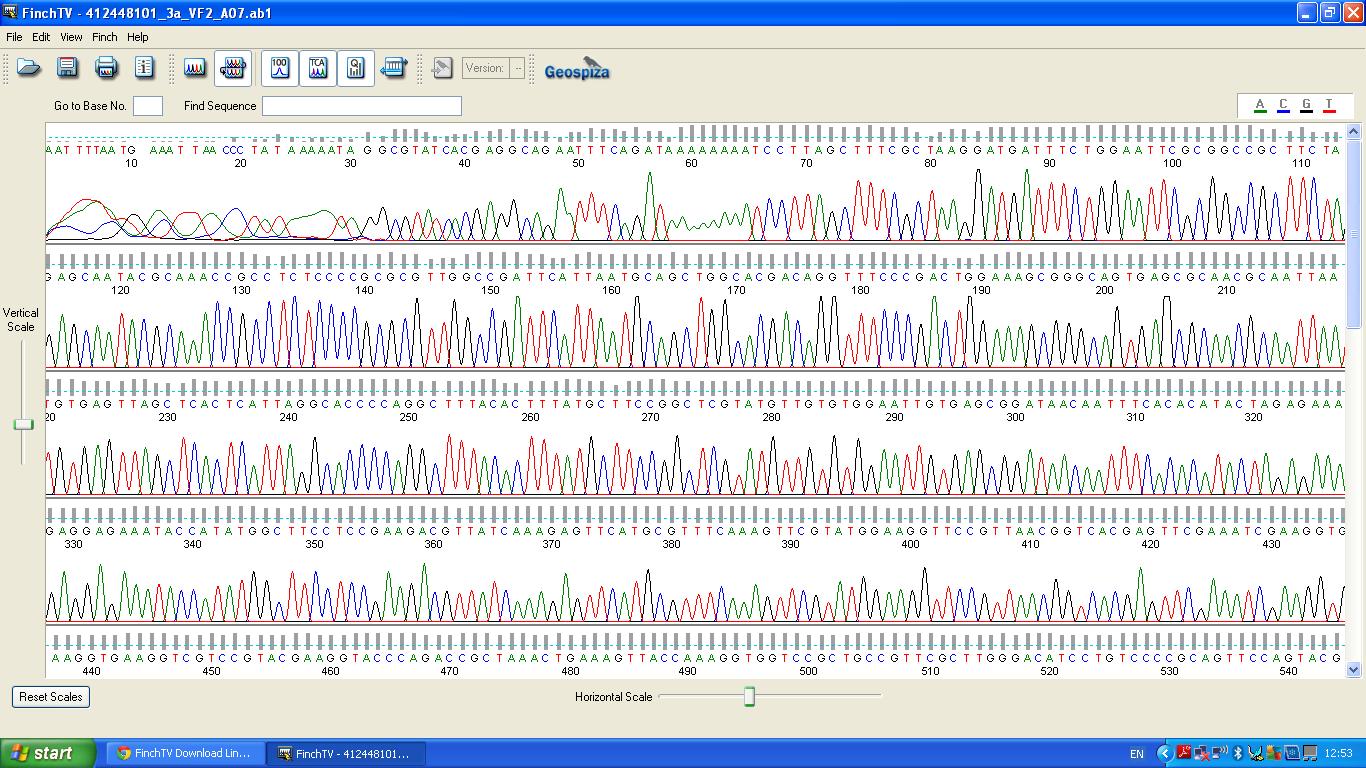
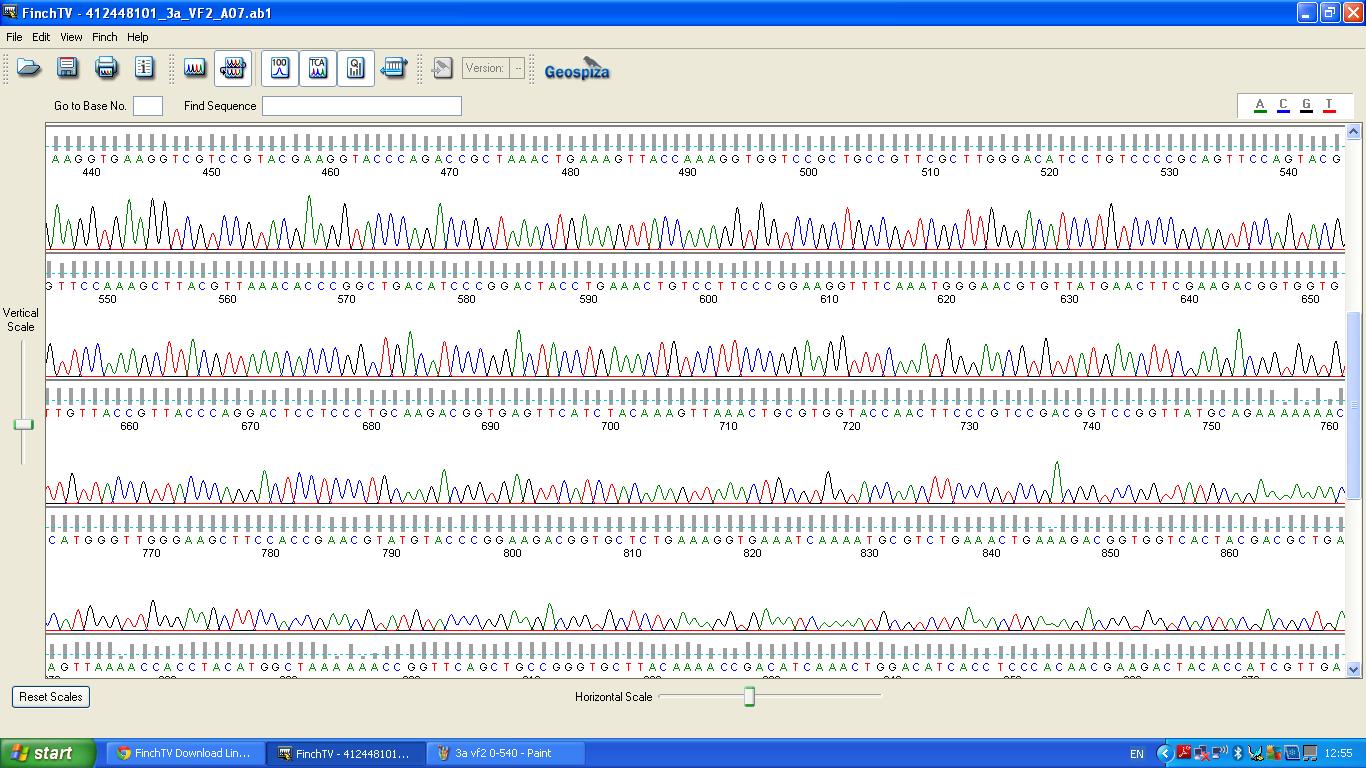
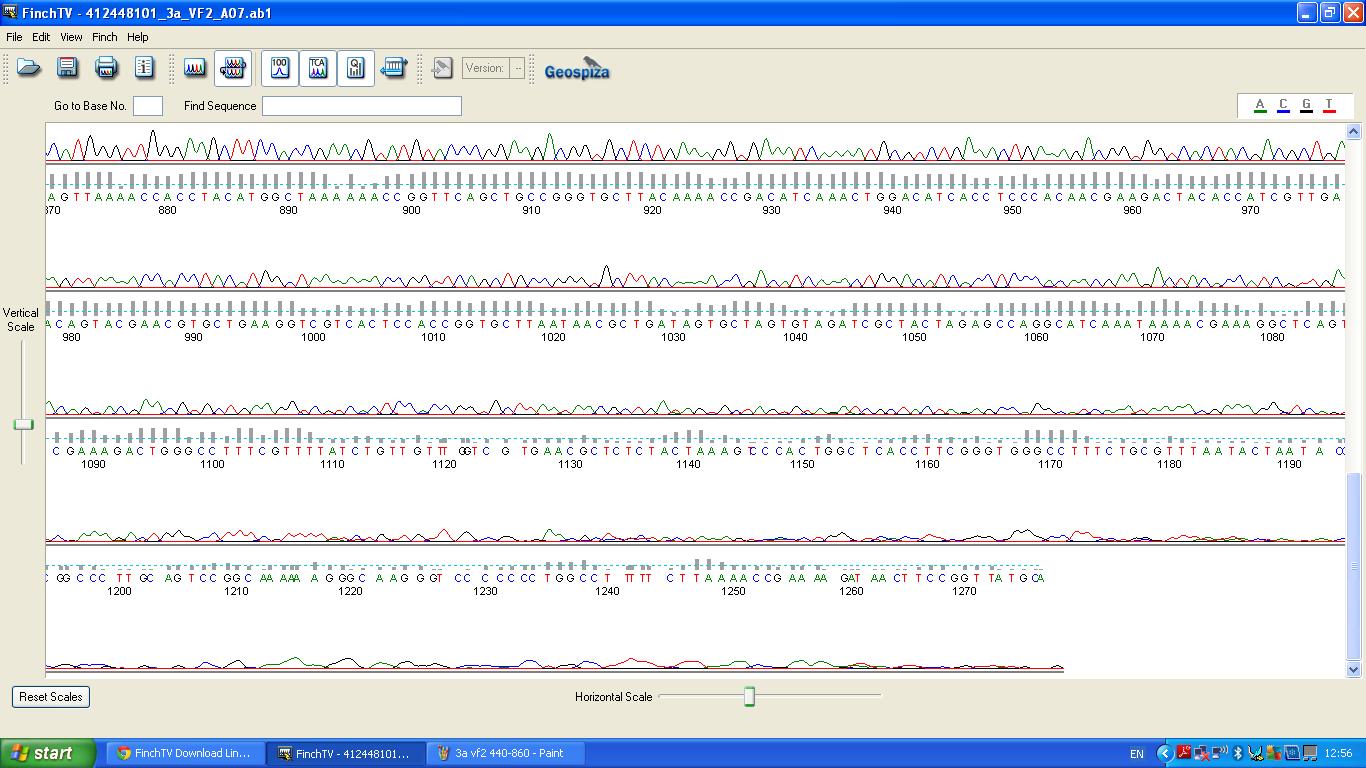
Characterisation of this biobrick involved sequencing, restriction digests and BLAST analysis. | Characterisation of this biobrick involved sequencing, restriction digests and BLAST analysis. | ||
| Line 27: | Line 37: | ||
===BLAST Analysis=== | ===BLAST Analysis=== | ||
The data we recieved back from the sequencing company was aligned using BLAST with the expected DNA sequence ''fig.2'' | The data we recieved back from the sequencing company was aligned using BLAST with the expected DNA sequence ''fig.2'' | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 11:22, 17 September 2013
AntG Promoter + RFP Coding Device
Team NRP-UEA_Norwich 2013 created this part using biobricks BBa_K1041000 and BBa_K1041001. These biobricks both contain a Nde1 site after their promoter sequence, enabling a restriction digest to be performed. The RFP coding gene was excised from BBa_K1041000 and ligated in front of the AntG promoter of BBa_K1041001 to create a new biobrick.
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 683
Illegal AgeI site found at 795 - 1000COMPATIBLE WITH RFC[1000]
Characterisation
Characterisation of this biobrick involved sequencing, restriction digests and BLAST analysis.
Sequencing
The biobrick was sent off to a company for sequencing.
BLAST Analysis
The data we recieved back from the sequencing company was aligned using BLAST with the expected DNA sequence fig.2