Difference between revisions of "Help:2013 DNA Distribution"
m |
m |
||
| Line 38: | Line 38: | ||
==What's included in the 2013 Distribution== | ==What's included in the 2013 Distribution== | ||
| − | [[Image:2013_distribution.jpeg|left]] | + | <div style="float: left;">[[Image:2013_distribution.jpeg|left]]</div> |
| + | <div style="float: left; margin-left: 20px; width: 400px;"> | ||
<b>Your Spring 2013 Distribution contains the following:</b><br /> | <b>Your Spring 2013 Distribution contains the following:</b><br /> | ||
[[Image:Green_BulletPt.png|link=]] DNA Distribution Kit Plates 1 - 4<br /> | [[Image:Green_BulletPt.png|link=]] DNA Distribution Kit Plates 1 - 4<br /> | ||
| Line 48: | Line 49: | ||
If there is an issue with your distribution kit, please send us an email to ''hq (at) igem . org'' | If there is an issue with your distribution kit, please send us an email to ''hq (at) igem . org'' | ||
| + | </div> | ||
<div style="clear: both;"></div> | <div style="clear: both;"></div> | ||
| Line 62: | Line 64: | ||
The 2013 Distribution Kit also includes a set of four linearized plasmid backbones: pSB1A3, pSB1C3, pSB1T3, and pSB1K3.m1. These plasmid backbones have been prepared via PCR and purified. Prior to ligation the linearized backbones will need to be digested with with EcoRI, PstI and DpnI (DpnI is optional: to cut up original template DNA used to create linearized plasmid backbone), leaving two ends ready to be ligated to a Biobrick™ part. All 2013 submissions will need to be in pSB1C3, so we recommend using our pSB1C3 linearized backbone for that purpose. | The 2013 Distribution Kit also includes a set of four linearized plasmid backbones: pSB1A3, pSB1C3, pSB1T3, and pSB1K3.m1. These plasmid backbones have been prepared via PCR and purified. Prior to ligation the linearized backbones will need to be digested with with EcoRI, PstI and DpnI (DpnI is optional: to cut up original template DNA used to create linearized plasmid backbone), leaving two ends ready to be ligated to a Biobrick™ part. All 2013 submissions will need to be in pSB1C3, so we recommend using our pSB1C3 linearized backbone for that purpose. | ||
| − | + | See the [[Help:Protocols/Linearized Plasmid Backbones | Linearized Plasmid Backbone protocol page]] for more in-depth instructions on how to use and make your own linearized plasmid backbones. | |
| Line 112: | Line 114: | ||
===DNA Kit Plate Instructions=== | ===DNA Kit Plate Instructions=== | ||
| − | To use the DNA in the Distribution Kit | + | <b>Before you use the DNA in the Distribution Kit Plates, be sure to test the efficiency of your competent cells with the Transformation Efficiency Test Kit.</b><br /> |
| − | :# With a pipette tip, punch a hole through the foil cover into the corresponding well of the | + | |
| + | To use the DNA in the Distribution Kit, follow these instructions:<br /> | ||
| + | ''Note: There is an estimated 2-3ng of DNA in each well, following this protocol, assume that you are transforming with 200-300pg/ul'' | ||
| + | |||
| + | :# With a pipette tip, punch a hole through the foil cover into the corresponding well of the part that you want. [https://parts.igem.org/Help:Spring_2011_DNA_distribution#DNA_Kit_Plate_Orientation Make sure you have properly oriented the plate]. We recommend that you do not remove the foil cover, as it could lead to cross contamination between the wells. | ||
:# Pipette 10uL of dH2O (distilled water) into the well. Pipette up and down a few times and let sit for 5 minutes to make sure the dried DNA is fully resuspended. We recommend that you do not use TE to resuspend the dried DNA. | :# Pipette 10uL of dH2O (distilled water) into the well. Pipette up and down a few times and let sit for 5 minutes to make sure the dried DNA is fully resuspended. We recommend that you do not use TE to resuspend the dried DNA. | ||
| − | :# [[Help:Transformation_Protocol|Transform]] | + | :# [[Help:Transformation_Protocol|Transform]] 1ul of the resuspended DNA into your desired competent cells, plate your transformation with the appropriate antibiotic* and grow overnight. |
| − | :#Pick a single colony and inoculate broth (again, with the correct antibiotic) and grow for 16 hours. | + | :# Pick a single colony and inoculate broth (again, with the correct antibiotic) and grow for 16 hours. |
| − | :#Use the resulting culture to [http://openwetware.org/wiki/Miniprep/Kit-free_high-throughput_protocol miniprep] the DNA AND make your own glycerol stock (for further instruction on making a glycerol see [http://openwetware.org/wiki/Endy:Making_a_long_term_stock_of_bacteria this page]). We recommend using the miniprepped DNA to run QC tests, such as restriction digests and sequencing. | + | :# Use the resulting culture to [http://openwetware.org/wiki/Miniprep/Kit-free_high-throughput_protocol miniprep] the DNA AND make your own glycerol stock (for further instruction on making a glycerol see [http://openwetware.org/wiki/Endy:Making_a_long_term_stock_of_bacteria this page]). We recommend using the miniprepped DNA to run QC tests, such as restriction digests and sequencing. |
''* To know which antibiotics to use, look at the plasmid that the part is in. The [https://parts.igem.org/wiki/index.php/Help:Plasmids/Nomenclature naming scheme] for plasmids is specifically designed to indicate antibiotic resistance.'' | ''* To know which antibiotics to use, look at the plasmid that the part is in. The [https://parts.igem.org/wiki/index.php/Help:Plasmids/Nomenclature naming scheme] for plasmids is specifically designed to indicate antibiotic resistance.'' | ||
Revision as of 15:40, 31 May 2013
| Template:HelpPage/MiniMap | ||
 |
Distribution Kits |
|
| The DNA Distribution has been entirely revamped this year! Whether you’re new to iGEM and the Registry of Standard Biological Parts or an experienced participant, please make sure to read through the Distribution Handbook. | ||
The 2013 Distribution contains over 1000 part samples as dried (miniprepped) DNA, with each sample QC tested through sequencing, AB test plates, and restriction digests. While there is not enough DNA for assembly, you will be able to transform the DNA into cells and then make your own glycerol stocks of any part you wish.
If you're new to the Registry, iGEM, or synthetic biology, you'll want to read through our Help pages before you get started. The Learn section is the best place to start, and will take you through the basics.
Otherwise, read on!
Changes to the 2013 DNA Distribution
We’ve made some important changes to this year’s DNA Distribution!
![]() In making this distribution, we selected only parts that had their sequence’s fully confirmed or ends confirmed (long parts).
In making this distribution, we selected only parts that had their sequence’s fully confirmed or ends confirmed (long parts).
![]() All parts in Kit Plates 1 to 4 are in pSB1C3, the shipping standard plasmid backbone for the iGEM Registry.
All parts in Kit Plates 1 to 4 are in pSB1C3, the shipping standard plasmid backbone for the iGEM Registry.
![]() We’ve included the Transformation Efficiency Kit, so that you can test the transformation efficiency of your competent cells. Learn more...
We’ve included the Transformation Efficiency Kit, so that you can test the transformation efficiency of your competent cells. Learn more...
What's included in the 2013 Distribution
Your Spring 2013 Distribution contains the following:
![]() DNA Distribution Kit Plates 1 - 4
DNA Distribution Kit Plates 1 - 4
![]() DNA Distribution Kit Plate 5 (Supplemental Plate)
DNA Distribution Kit Plate 5 (Supplemental Plate)
![]() Linearized Plasmid Backbones
Linearized Plasmid Backbones
![]() Transformation Efficiency Kit
Transformation Efficiency Kit
![]() iGEM Stickers and Pins! (iGEM teams only)
iGEM Stickers and Pins! (iGEM teams only)
If there is an issue with your distribution kit, please send us an email to hq (at) igem . org
Transformation Efficiency Kit
Whether you’re purchasing competent cells or making your own in the lab, you should test their efficiency before you use the parts in the DNA Distribution Kit Plates. The Transformation Efficiency Kit is a standardized way to test and calculate the efficiency of your competent cells. If you have issues with using the parts in the Distribution Kit, the first thing we’ll need to know is the efficiency of your competent cells.
See the Transformation Efficiency Kit page for more in-depth instructions.
Linearized Plasmid Backbones
The 2013 Distribution Kit also includes a set of four linearized plasmid backbones: pSB1A3, pSB1C3, pSB1T3, and pSB1K3.m1. These plasmid backbones have been prepared via PCR and purified. Prior to ligation the linearized backbones will need to be digested with with EcoRI, PstI and DpnI (DpnI is optional: to cut up original template DNA used to create linearized plasmid backbone), leaving two ends ready to be ligated to a Biobrick™ part. All 2013 submissions will need to be in pSB1C3, so we recommend using our pSB1C3 linearized backbone for that purpose.
See the Linearized Plasmid Backbone protocol page for more in-depth instructions on how to use and make your own linearized plasmid backbones.
Getting Started
Locating a Part in the Distribution
Before using the DNA plates, you should search the Registry for useful parts, which will also tell you if it has an available sample, its location (if they're in your 2013 Distribution), requirements, quality control (if they're correct), etc.
Searching
- The Catalog of Parts and Devices will allow you to browse parts and devices by various criteria, including function, chassis, standard, etc.
- The Search Tools will let you search by text or part name <more>.
- It is also possible to see the contents of the Kit Plates in their entirety. You can look at the DNA Repositories section on the main page.
- Click on 2013 Distribution to see the contents of all of your kit plates.
- Click on the 2013 Kit Plate of your choice, which will list all parts by their part name (BBa_..) in a plate along with their quality control information. Or you can click on the small part diagram below each Kit Plate link: "See a summary of the parts in this plate."
- These options will show you what is in each well of your plate, however they are not the best way to find specific parts you would like to use.
Part Main Page
When you find a part you can learn all about it by going through its main page and sub-pages.
Availability
If you find a part that you would like to use, you need to make sure that a sample of the part is actually available. There are many parts in the Registry that people are still working on, or decided not to continue working on anymore, therefore we never received or do not yet have the physical DNA for them. This of course means that the DNA is not available in the Spring 2013 DNA Distribution. The simplest way to tell whether the part has an available sample is to look at the top right of the part's Main Page. If the part has an available sample the top part of the box will be green and say "DNA available."
Requesting a part
We've taken care to create a very well-rounded DNA Distribution this spring, however, should you find the part, or parts, that you require are not available in the distribution but available in the Registry's Repository, send us an email (hq [AT] igem [DOT] org) in order to request a sample. Just include the part name, the plasmid it's located in, and the source, and we'll send it out to you. For more information please see the Requesting Parts page
Using the online QC resources
Here at Registry, we want everyone to take a look at the results of the quality control measures we've taken this year and previous years, in order to make an informed decision when choosing to use a part. We've made sure to update the online repository for the Spring 2013 distribution with our quality control results.
The best way to use our quality control information is to use it on a part by part basis. As you design your project, make sure to check every part that you're interested in for its QC data. After searching for a part in the registry and arriving at its main page, click on the Get This Part link which will take you to the section listing various quality control information for a samples of that part and its location within the registry.
Using the Spring 2013 DNA Distribution
Storage
Since the distribution kit plates are comprised of dried DNA, they are quite stable for storage at room temperature. However once the DNA is resuspended in any of the wells, we recommend either storing the kit plate with its plastic cover in a -20C freezer, or aspirating the rest of the resuspended DNA from the well and keeping it separately in a -20C freezer.
The linearized plasmid backbones (25ng/ul at 50ul) should be stored at 4C or lower.
DNA Kit Plate Orientation
Locating Your Part from iGEM Videos.
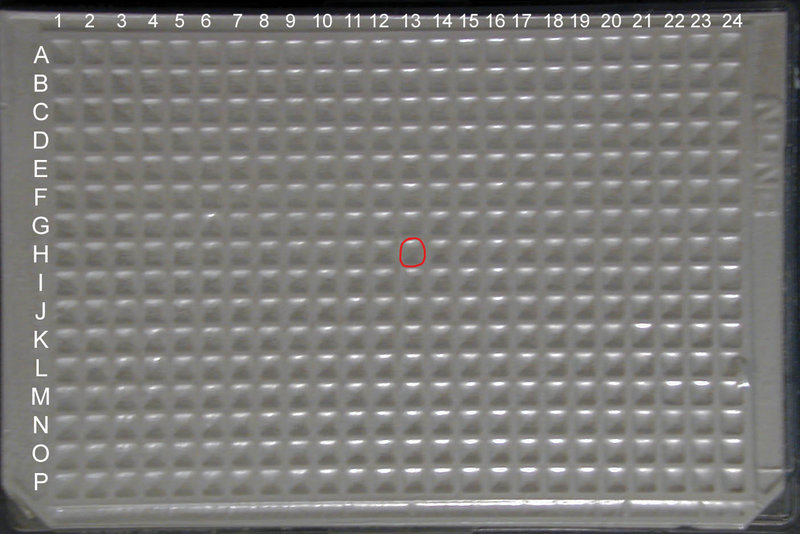
The foil covers on each 384 well kit plate can be easily punched through with a pipette tip into the well of your choice. Unfortunately, the foil cover will also obscure both column and well markings. You can still find your part by correctly orienting the plate using the two notched corners as markers: well A1 is located at the upper left corner of the plate when the long side of the plate with the notched corners is considered the bottom.
Once you know the location of your part, you will want to count across the plate starting with Column 1 until you get to Column 13 and down the plate starting with Row A until you get to Row H.
Make sure that the two notched corners of the plate are oriented at the BOTTOM of the plate (see the "top view" image on the right for correct orientation)
DNA Kit Plate Instructions
Before you use the DNA in the Distribution Kit Plates, be sure to test the efficiency of your competent cells with the Transformation Efficiency Test Kit.
To use the DNA in the Distribution Kit, follow these instructions:
Note: There is an estimated 2-3ng of DNA in each well, following this protocol, assume that you are transforming with 200-300pg/ul
- With a pipette tip, punch a hole through the foil cover into the corresponding well of the part that you want. Make sure you have properly oriented the plate. We recommend that you do not remove the foil cover, as it could lead to cross contamination between the wells.
- Pipette 10uL of dH2O (distilled water) into the well. Pipette up and down a few times and let sit for 5 minutes to make sure the dried DNA is fully resuspended. We recommend that you do not use TE to resuspend the dried DNA.
- Transform 1ul of the resuspended DNA into your desired competent cells, plate your transformation with the appropriate antibiotic* and grow overnight.
- Pick a single colony and inoculate broth (again, with the correct antibiotic) and grow for 16 hours.
- Use the resulting culture to [http://openwetware.org/wiki/Miniprep/Kit-free_high-throughput_protocol miniprep] the DNA AND make your own glycerol stock (for further instruction on making a glycerol see [http://openwetware.org/wiki/Endy:Making_a_long_term_stock_of_bacteria this page]). We recommend using the miniprepped DNA to run QC tests, such as restriction digests and sequencing.
* To know which antibiotics to use, look at the plasmid that the part is in. The naming scheme for plasmids is specifically designed to indicate antibiotic resistance.
Note: There is not enough DNA in each well to perform anything but transformations
Get & Use...