Difference between revisions of "Part:BBa K658016"
(rollbackEdits.php mass rollback) |
|||
| (15 intermediate revisions by 4 users not shown) | |||
| Line 16: | Line 16: | ||
[[Image:XMU_China_3_forframe.jpg|left|Figure 3: the sequence of promoter lux pR.|frame|Figure 3: the sequence of promoter lux pR.]] | [[Image:XMU_China_3_forframe.jpg|left|Figure 3: the sequence of promoter lux pR.|frame|Figure 3: the sequence of promoter lux pR.]] | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/4/41/XMU_China_block.jpg"> | ||
| + | </html> | ||
| + | |||
On the basis of the nucleotide sequence of the lux pR promoter, the 20 base pair inverted repeat ACCTGTAGGA TCGTACAGGT might consititude a protein binding site. And we also learned that mutagenesis at position 3 and position 5 might cause dramatic change on the expression of downstream gene. Therefore, we first generated 3 mutants (BBa_K658006 BBa_K658007 BBa_K658008) of the promoter lux pR by site-directed mutagenesis, then we tested their strength in this IR-GFP device by measuring fluorescence at steady state. | On the basis of the nucleotide sequence of the lux pR promoter, the 20 base pair inverted repeat ACCTGTAGGA TCGTACAGGT might consititude a protein binding site. And we also learned that mutagenesis at position 3 and position 5 might cause dramatic change on the expression of downstream gene. Therefore, we first generated 3 mutants (BBa_K658006 BBa_K658007 BBa_K658008) of the promoter lux pR by site-directed mutagenesis, then we tested their strength in this IR-GFP device by measuring fluorescence at steady state. | ||
| Line 24: | Line 30: | ||
The results are shown in following figures: | The results are shown in following figures: | ||
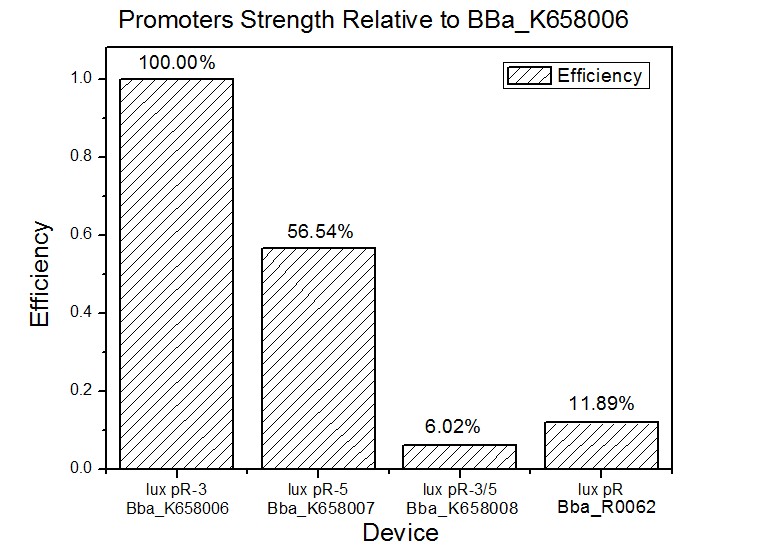
| − | [[Image: | + | [[Image:XMU_China_4beta.jpg|left|Figure 4 Promoters strength relative to lux pR-3 (BBa_K658006)|frame|Figure 4 Promoters strength relative to lux pR-3 (BBa_K658006).]] |
| − | [[Image: | + | [[Image:XMU_China_5beta.jpg|left|Figure 5 Efficiency of promoter lux pR (BBa_R0062) and its three mutants |
| + | |frame|Figure 5 Efficiency of promoter lux pR (BBa_R0062) and its three mutants.]] | ||
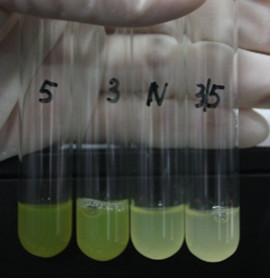
| − | [[Image: | + | [[Image:XMU_China_6beta.jpg|left|Figure 6: Fluorescence of four IR-GFP devices at 20h. 5, 3, 3/5, N represent for IR-3-GFP (BBa_K658017), IR-5-GFP (BBa_K658018), IR-3/5-GFP (BBa_K658019) and IR-GFP (BBa_K658016) respectively|frame|Figure 6: Fluorescence of four IR-GFP devices at 20h. 5, 3, 3/5, N represent for IR-3-GFP (BBa_K658017), IR-5-GFP (BBa_K658018), IR-3/5-GFP (BBa_K658019) and IR-GFP (BBa_K658016) respectively.]] |
<html> | <html> | ||
| Line 36: | Line 43: | ||
Figure 4, figure 5 and figure 6 illustrate that mutated promoters lux pR-3 (BBa_K658006) and lux R-5 (BBa_K658007) dramatically increased the fluorescence intensity at steady state compared with wild type promoter lux pR (R0062), while mutated promoter lux pR-3/5 (BBa_K658008) gave an even weaker expression of GFP than promoter lux pR (R0062). It might be explained that the mutagenesis at position 3 and position 5 of the sequence of lux pR (R0062) changed the binding strength between promoter lux pR and protein luxR. | Figure 4, figure 5 and figure 6 illustrate that mutated promoters lux pR-3 (BBa_K658006) and lux R-5 (BBa_K658007) dramatically increased the fluorescence intensity at steady state compared with wild type promoter lux pR (R0062), while mutated promoter lux pR-3/5 (BBa_K658008) gave an even weaker expression of GFP than promoter lux pR (R0062). It might be explained that the mutagenesis at position 3 and position 5 of the sequence of lux pR (R0062) changed the binding strength between promoter lux pR and protein luxR. | ||
| + | As is shown in figure 5, promoter lux pR-3 has the highest strength of the four. Mutation at position 3 might lower the threshold for the binding reaction between LuxR/AHL protein complex and promoter lux pR, which starts the Quorum Sensing system at a relatively earlier period with a lower cell density compared with circuits regulated by wild type promoter lux pR (BBa_R0062). The earlier the QS system is started, the more GFP might be produced, leading to a higher fluorescence intensity at steady state. | ||
== Method == | == Method == | ||
| Line 42: | Line 50: | ||
Using the Procedure for GenEluteTM Plasmid Miniprep Kit | Using the Procedure for GenEluteTM Plasmid Miniprep Kit | ||
| − | + | •Collect 1-5 mL bacterium fluid in 1.5 mL centrifuge tube,and centrifuge fluid at 12000 r/min | |
| − | + | •Resuspend cells. Discard the supernatant and completely resuspend the bacterial pellet with 250 µl of the Resuspension Solution | |
| − | + | •Lyse cells. Lyse the resuspended cells by adding 250 µl of the Lysis Solution | |
| − | + | •Neutralize. Precipitate the cell debris by adding 350 µl of the Neutralization/Binding Solution, and centrifuge fluid at 12000r/min | |
| − | + | •Load cleared lysate. Transfer the supernatant from step 4 to the spin column. Centrifuge at 12000r/min for 1 minute, and then discard the filtrate | |
| − | + | •Optional wash. Add 500 µl of the Optional Wash Solution to the column. Centrifuge at 12000 r/min for 1 minute. Discard the filtrate | |
| − | + | •Wash column. Add 500 µl of the diluted Wash Solution to the column. Centrifuge at 12000r/min for 1 minute. | |
| − | + | •Elute DNA. Transfer the column to a new collection tube. Add 50~100 µl of Eluent Solution to the column. Centrifuge | |
at 12000 r/min for 1 minute. The DNA is now present in the filtrate and is ready for immediate use or storage at -20℃ | at 12000 r/min for 1 minute. The DNA is now present in the filtrate and is ready for immediate use or storage at -20℃ | ||
| Line 62: | Line 70: | ||
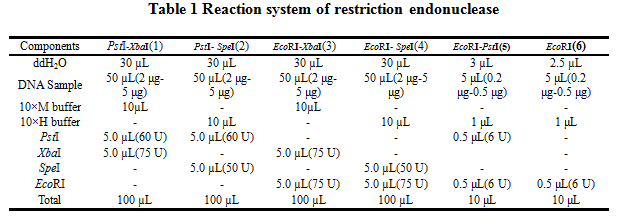
'''2.Reaction system of restriction endonuclease''' | '''2.Reaction system of restriction endonuclease''' | ||
| − | [[Image:XMU_China_7.jpg|left | + | [[Image:XMU_China_7.jpg|left]] |
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/4/41/XMU_China_block.jpg"> | ||
| + | </html> | ||
| Line 69: | Line 80: | ||
'''3. Standard BioBrick Assembly''' | '''3. Standard BioBrick Assembly''' | ||
| − | + | •Digestion of insert: 2 μg~5 μg DNA / 100 µL volume, 10x H buffer, EcoRI, SpeI. Digestion and inactivation. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel, try to avoid staining or exposure to ultraviolet light of the insert. | |
| − | + | •Digestion of vector: 2 μg~5 μg DNA / 100 µL volume, 10 x M buffer, EcoRI, XbaI. Digestion and inactivation. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel, try to avoid staining or exposure to ultraviolet light of the insert. | |
'''4. Suffix Insertion''' | '''4. Suffix Insertion''' | ||
| − | + | •Digestion of insert: 2 μg~5 μg DNA / 100 µL volume, 10x M buffer, XbaI, PstI. Digestion and inactivation. Clean up the insert. | |
| − | + | •Digestion of vector : 2 μg~5 μg DNA / 100 µL volume, 10x H buffer, SpeI, PstI. Digestion and inactivation. Clean up the vector. | |
'''5. Ligation''' | '''5. Ligation''' | ||
| − | + | •After digestion and clean-up, the next step is ligation. Overnight ligation at 16°C. Table 2 is the system of ligation. | |
'''6. Transformation''' | '''6. Transformation''' | ||
| − | + | •Preparation of competent E.coli cells | |
| − | + | •Add 10 µL plasmid to 100 µl competent cells in centrifuge tube | |
| − | + | •Store tube on ice for 20-30 minutes | |
| − | + | •Water bath for 90s at 42℃ | |
| − | + | •Put the tube on ice for 1-2min | |
| − | + | •Add 790 µL LB,and cultivation for 1h at 37 ℃,then plate on selective LB-Medium. | |
'''7. Restriction analysis''' | '''7. Restriction analysis''' | ||
| − | + | •Pick one colony with a sterile tip and cultivation in 20ml LB for overnight at 37 ℃ | |
| − | + | •Isolation of Plasmid | |
| − | + | •Digest BioBrick,the system of Restriction analysis refer to table1 | |
| − | + | •Gel electrophoresis:add 2 µL loading buffer to digestion mixture. An agarose concentration is 1 %. | |
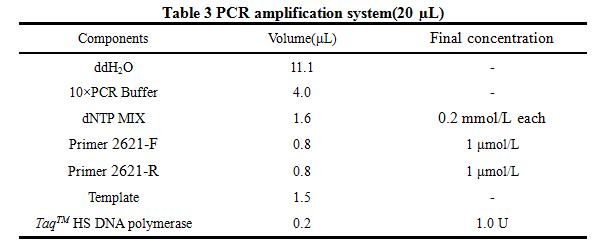
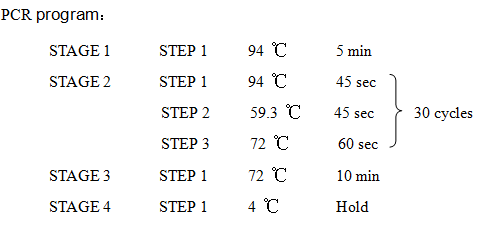
'''8. PCR''' | '''8. PCR''' | ||
| Line 119: | Line 130: | ||
2621-R:5'-AAAACTGCAGCGGCCGCTACTA-3' | 2621-R:5'-AAAACTGCAGCGGCCGCTACTA-3' | ||
| − | [[Image:XMU_China_8.jpg|left | + | [[Image:XMU_China_8.jpg|left]] |
| − | [[Image:XMU_China_9.jpg|left | + | [[Image:XMU_China_9.jpg|left]] |
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/4/41/XMU_China_block.jpg"> | ||
| + | </html> | ||
| Line 127: | Line 141: | ||
'''9. Cell growth''' | '''9. Cell growth''' | ||
| − | + | •100µL suspension was inoculated from a Glycerin tube into 20ml fresh LB and incubated overnight at 37℃ and 250 r.p.m. | |
| − | + | •100µL suspension was inoculated again from step1 into 50ml fresh LB and incubated at 37℃ and 250r.p.m | |
| − | + | •IPTG was added when A600≈0.6-0.8 | |
| − | + | •1 ml suspension was taken on every sample taken time. 3 samples were taken in each time. | |
| − | + | •Diluted each sample to 10-6(Sometimes 10-5),and then plate on selective LB-Medium. | |
| − | + | •After 12h, count the number of CFU on the plate on different time point and then draw the cell growth curve. | |
[[Image:XMU_China_10.jpg|left|frame]] | [[Image:XMU_China_10.jpg|left|frame]] | ||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/4/41/XMU_China_block.jpg"> | ||
| + | </html> | ||
'''10. Determining fluorescence intensity''' | '''10. Determining fluorescence intensity''' | ||
| − | + | •Add IPTG when A600 0.6~0.8. | |
| − | + | •Cool the culture 10 minutes on ice. | |
| − | + | •Centrifuge at 6000rpm. Wash it with pre-cooled PBS buffer. | |
| − | + | •Use fluorescence spectrophotometer tomeasure the fluorescenceof GFP: | |
| − | + | •Before measuring, dilute the bacteria with PBS buffer so that it can be within the measuring #range. Set excitation wavelength 491 nm, emission wavelength 511 nm. | |
| − | + | •Transfer the measured bacteria in a new centrifuge tube and measure the OD of the bacteria. | |
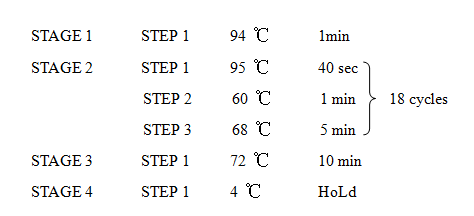
'''11. Site Directed Mutagenesis''' | '''11. Site Directed Mutagenesis''' | ||
| − | [[Image:XMU_China_11.jpg|left | + | [[Image:XMU_China_11.jpg|left]] |
| − | [[Image:XMU_China_12.jpg|left | + | [[Image:XMU_China_12.jpg|left]] |
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/4/41/XMU_China_block.jpg"> | ||
| + | </html> | ||
•Digest the template plasmid by adding 1 µL of DpnI and incubate for 1-2 h | •Digest the template plasmid by adding 1 µL of DpnI and incubate for 1-2 h | ||
| Line 170: | Line 190: | ||
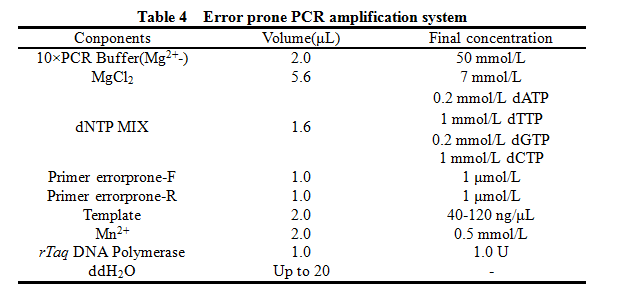
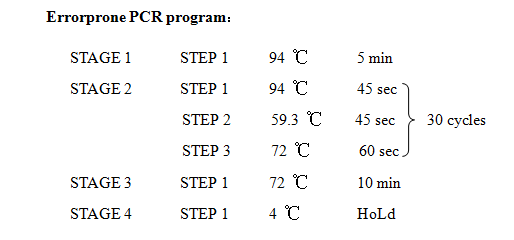
'''12. Error prone PCR''' | '''12. Error prone PCR''' | ||
| − | [[Image:XMU_China_13.jpg|left| | + | [[Image:XMU_China_13.jpg|left]] |
| + | |||
| + | [[Image:XMU_China_14.jpg|left]] | ||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/4/41/XMU_China_block.jpg"> | ||
| + | </html> | ||
| + | |||
| + | ==reference== | ||
| + | [1]Baldwin T, Devine JH, Heckel, RC, Lin, JW, Shadel GS. The complete nucleotide sequence of the lux regulon of Vibrio fischeri and the luxABN region of Photobacterium leiognathi and the mechanism of control of bacterial bioluminescence[J]. Journal of Bioluminescence and Chemiluminescence, 1989, 4(1): 326-341. | ||
| + | |||
| + | [2]Philippe B,Martine C.Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes[J].Molecular Biology,1992,226(3):735-745. | ||
| + | |||
| + | [3]Devine J.H,Shadel G.S,Baldwin T.O. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proceedings of the National Academy of Sciences.1989.86(15):5688-5692. | ||
| + | |||
| + | [4] http://www.uniprot.org/uniprot/P12747 | ||
| + | |||
| + | [5]Miki T,Yoshioka K,Horiuchi T. Control of cell division by sex factor F in Escherichia coli. I. The 42.84-43.6 F segment couples cell division of the host bacteria with replication of plasmid DNA. Molecular Biology,1984,177(4):605-625. | ||
| + | |||
| + | [6] http://en.wikipedia.org/wiki/N-Acyl_homoserine_lactone | ||
| + | |||
| + | [7] Hanzelka BL, Greenberg E. Quorum sensing in Vibrio fischeri: evidence that S-adenosyl methionine is the amino acid substrate for autoinducer synthesis[J]. Journal of Bacteriology, 1996, 178(17): 5291-5294. | ||
| + | |||
| + | [8] Urbanowski M, Lostroh C, Greenberg E. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein[J]. Journal of Bacteriology, 2004, 186(3): 631-637. | ||
| + | |||
| + | [9] Hansen LH, Knudsen S, Sorensen SJ . The effect of the lacY gene on the induction of IPTG inducible promoters, studied in Escherichia coli and Pseudomonas fluorescens. Curr microbiol,1998, 36 (6): 341–345. | ||
| + | |||
| + | [10]Bernard P,Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. Molecular Biology,1992,226:735-745. | ||
| − | |||
Latest revision as of 16:18, 10 May 2013
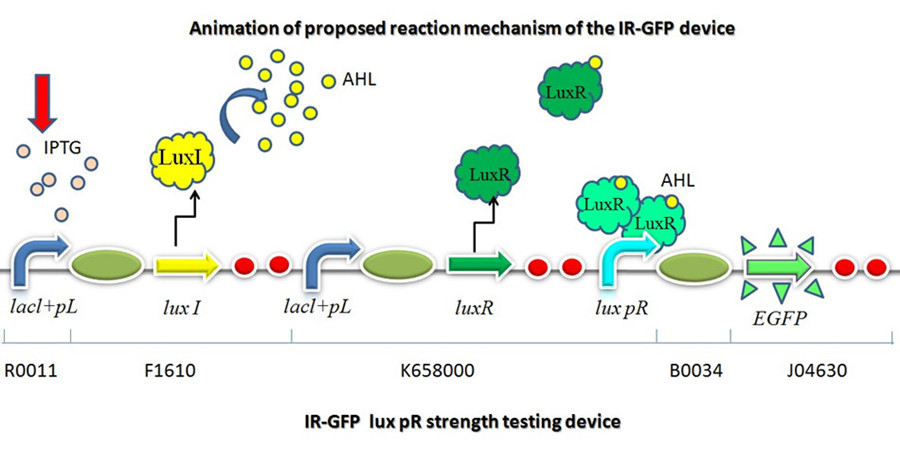
LuxI->LuxR->lux pR->GFP
Lux pR strength testing device
This part is designed to test the strength of promoters lux pR(BBa_R0062) and its 3 mutants lux pR-3 (BBa_K658006), lux pR-5(BBa_K658007), lux pR-3/5(BBa_K658008) in our project. If promoter lacl+pL(BBa_R0011) is induced by isopropyl-b-D-thiogalactopyranoside (IPTG), this device will be switched on. At sufficiently high cell density, this device produces greenish tint visible by naked eye. By measuring florescent intensities at steady state of the cell growth for these four IR-GFP devices, the strength of a promoter lux pR could be defined.
Description
This device is made up of three subparts: a LuxI producer, a Lux R producer and a EGFP reporter. The luxI producer and luxR producer in this device work as a 3OC6HSL sender and receiver device driven by lacl+pL(R0011). We constructed the part which expresses LuxI and LuxR upon induction by isopropyl-b-D-thiogalactopyranoside (IPTG). There are two regulatory genes involved in quorum sensing, the I and R genes. The I gene directs the synthesis of an acyl-homoserine lactone (AHL) signal molecule termed the autoinducer. The R gene codes for a transcription factor that is responsive to the N-acyl HSL signal. Cells are permeable to this signal, and thus high cell densities are required to achieve a critical concentration of the autoinducer required to bind the luxR product, which will combine with lux pR and active the EGFP to transcribe.

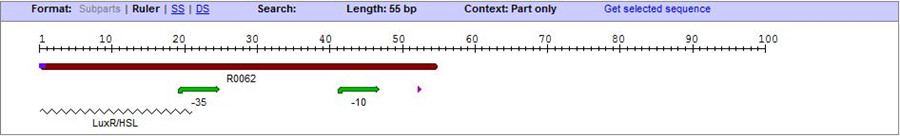
On the basis of the nucleotide sequence of the lux pR promoter, the 20 base pair inverted repeat ACCTGTAGGA TCGTACAGGT might consititude a protein binding site. And we also learned that mutagenesis at position 3 and position 5 might cause dramatic change on the expression of downstream gene. Therefore, we first generated 3 mutants (BBa_K658006 BBa_K658007 BBa_K658008) of the promoter lux pR by site-directed mutagenesis, then we tested their strength in this IR-GFP device by measuring fluorescence at steady state.
Result
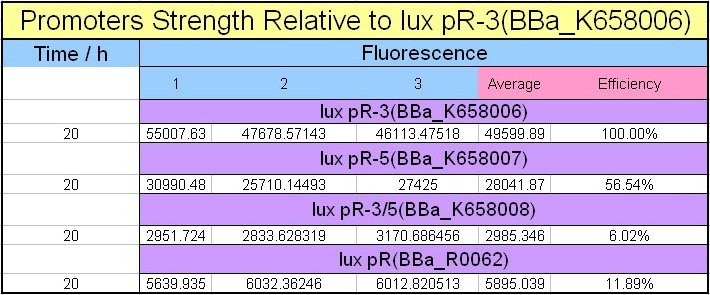
Four lux pR strength testing devices (BBa_K658016 BBa_K658017 BBa_K658018 BBa_K658019) were first cloned into plasmid pSB1A2 respectively, followed by transformation into E.coli strain BL21. Fluorescence was measured when cell growth reached a steady state (around 20h). The results are shown in following figures:

Figure 4, figure 5 and figure 6 illustrate that mutated promoters lux pR-3 (BBa_K658006) and lux R-5 (BBa_K658007) dramatically increased the fluorescence intensity at steady state compared with wild type promoter lux pR (R0062), while mutated promoter lux pR-3/5 (BBa_K658008) gave an even weaker expression of GFP than promoter lux pR (R0062). It might be explained that the mutagenesis at position 3 and position 5 of the sequence of lux pR (R0062) changed the binding strength between promoter lux pR and protein luxR.
As is shown in figure 5, promoter lux pR-3 has the highest strength of the four. Mutation at position 3 might lower the threshold for the binding reaction between LuxR/AHL protein complex and promoter lux pR, which starts the Quorum Sensing system at a relatively earlier period with a lower cell density compared with circuits regulated by wild type promoter lux pR (BBa_R0062). The earlier the QS system is started, the more GFP might be produced, leading to a higher fluorescence intensity at steady state.
Method
1.Isolation of Plasmid
Using the Procedure for GenEluteTM Plasmid Miniprep Kit
•Collect 1-5 mL bacterium fluid in 1.5 mL centrifuge tube,and centrifuge fluid at 12000 r/min
•Resuspend cells. Discard the supernatant and completely resuspend the bacterial pellet with 250 µl of the Resuspension Solution
•Lyse cells. Lyse the resuspended cells by adding 250 µl of the Lysis Solution
•Neutralize. Precipitate the cell debris by adding 350 µl of the Neutralization/Binding Solution, and centrifuge fluid at 12000r/min
•Load cleared lysate. Transfer the supernatant from step 4 to the spin column. Centrifuge at 12000r/min for 1 minute, and then discard the filtrate
•Optional wash. Add 500 µl of the Optional Wash Solution to the column. Centrifuge at 12000 r/min for 1 minute. Discard the filtrate
•Wash column. Add 500 µl of the diluted Wash Solution to the column. Centrifuge at 12000r/min for 1 minute.
•Elute DNA. Transfer the column to a new collection tube. Add 50~100 µl of Eluent Solution to the column. Centrifuge at 12000 r/min for 1 minute. The DNA is now present in the filtrate and is ready for immediate use or storage at -20℃
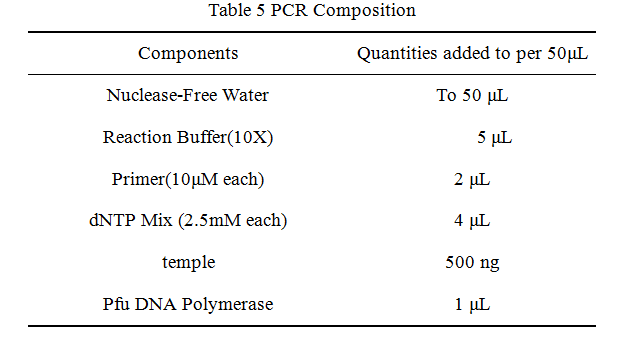
2.Reaction system of restriction endonuclease

System1、2、3 and 4 are used for Standard BioBrick Assembly .System 5 and 6 are used for Restriction analysis. Digestion of sample: at least 500 ng DNA / 10 µL volume. Digest for 4 h at 37 °C, afterwards inactivated by adding 10x loading buffer and standing for 10 min at room temperature.
3. Standard BioBrick Assembly
•Digestion of insert: 2 μg~5 μg DNA / 100 µL volume, 10x H buffer, EcoRI, SpeI. Digestion and inactivation. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel, try to avoid staining or exposure to ultraviolet light of the insert.
•Digestion of vector: 2 μg~5 μg DNA / 100 µL volume, 10 x M buffer, EcoRI, XbaI. Digestion and inactivation. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel, try to avoid staining or exposure to ultraviolet light of the insert.
4. Suffix Insertion
•Digestion of insert: 2 μg~5 μg DNA / 100 µL volume, 10x M buffer, XbaI, PstI. Digestion and inactivation. Clean up the insert.
•Digestion of vector : 2 μg~5 μg DNA / 100 µL volume, 10x H buffer, SpeI, PstI. Digestion and inactivation. Clean up the vector.
5. Ligation
•After digestion and clean-up, the next step is ligation. Overnight ligation at 16°C. Table 2 is the system of ligation.
6. Transformation
•Preparation of competent E.coli cells
•Add 10 µL plasmid to 100 µl competent cells in centrifuge tube
•Store tube on ice for 20-30 minutes
•Water bath for 90s at 42℃
•Put the tube on ice for 1-2min
•Add 790 µL LB,and cultivation for 1h at 37 ℃,then plate on selective LB-Medium.
7. Restriction analysis
•Pick one colony with a sterile tip and cultivation in 20ml LB for overnight at 37 ℃
•Isolation of Plasmid
•Digest BioBrick,the system of Restriction analysis refer to table1
•Gel electrophoresis:add 2 µL loading buffer to digestion mixture. An agarose concentration is 1 %.
8. PCR -RBS1.0-luxR-TT-lux pR-
Design Primers using the primer 5
Primer:
2621-F:5'-GCTCTAGAGAAAGAGGAGAAATACT-3'
2621-R:5'-AAAACTGCAGCGGCCGCTACTA-3'

9. Cell growth
•100µL suspension was inoculated from a Glycerin tube into 20ml fresh LB and incubated overnight at 37℃ and 250 r.p.m.
•100µL suspension was inoculated again from step1 into 50ml fresh LB and incubated at 37℃ and 250r.p.m
•IPTG was added when A600≈0.6-0.8
•1 ml suspension was taken on every sample taken time. 3 samples were taken in each time.
•Diluted each sample to 10-6(Sometimes 10-5),and then plate on selective LB-Medium.
•After 12h, count the number of CFU on the plate on different time point and then draw the cell growth curve.

10. Determining fluorescence intensity
•Add IPTG when A600 0.6~0.8.
•Cool the culture 10 minutes on ice.
•Centrifuge at 6000rpm. Wash it with pre-cooled PBS buffer.
•Use fluorescence spectrophotometer tomeasure the fluorescenceof GFP:
•Before measuring, dilute the bacteria with PBS buffer so that it can be within the measuring #range. Set excitation wavelength 491 nm, emission wavelength 511 nm.
•Transfer the measured bacteria in a new centrifuge tube and measure the OD of the bacteria.
11. Site Directed Mutagenesis

•Digest the template plasmid by adding 1 µL of DpnI and incubate for 1-2 h
•Transform 10 µL of t PCR product into competent E. coli cells
•Screen the transformants using restiction digest and sequencing
12. Error prone PCR

reference
[1]Baldwin T, Devine JH, Heckel, RC, Lin, JW, Shadel GS. The complete nucleotide sequence of the lux regulon of Vibrio fischeri and the luxABN region of Photobacterium leiognathi and the mechanism of control of bacterial bioluminescence[J]. Journal of Bioluminescence and Chemiluminescence, 1989, 4(1): 326-341.
[2]Philippe B,Martine C.Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes[J].Molecular Biology,1992,226(3):735-745.
[3]Devine J.H,Shadel G.S,Baldwin T.O. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proceedings of the National Academy of Sciences.1989.86(15):5688-5692.
[4] http://www.uniprot.org/uniprot/P12747
[5]Miki T,Yoshioka K,Horiuchi T. Control of cell division by sex factor F in Escherichia coli. I. The 42.84-43.6 F segment couples cell division of the host bacteria with replication of plasmid DNA. Molecular Biology,1984,177(4):605-625.
[6] http://en.wikipedia.org/wiki/N-Acyl_homoserine_lactone
[7] Hanzelka BL, Greenberg E. Quorum sensing in Vibrio fischeri: evidence that S-adenosyl methionine is the amino acid substrate for autoinducer synthesis[J]. Journal of Bacteriology, 1996, 178(17): 5291-5294.
[8] Urbanowski M, Lostroh C, Greenberg E. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein[J]. Journal of Bacteriology, 2004, 186(3): 631-637.
[9] Hansen LH, Knudsen S, Sorensen SJ . The effect of the lacY gene on the induction of IPTG inducible promoters, studied in Escherichia coli and Pseudomonas fluorescens. Curr microbiol,1998, 36 (6): 341–345.
[10]Bernard P,Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. Molecular Biology,1992,226:735-745.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 718
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1874
Illegal BsaI.rc site found at 2601