Difference between revisions of "Part:BBa K801042"
| Line 22: | Line 22: | ||
==== LexA based light-switchable-promoter system ==== | ==== LexA based light-switchable-promoter system ==== | ||
| − | + | For more information about the LexA based system, please see here: [parts.igem.org/wiki/index.php?title=Part:BBa_K801043 BBa_K801043] | |
| − | + | ||
| − | + | ||
=== Biosynthesis of phycocyanobilin === | === Biosynthesis of phycocyanobilin === | ||
Revision as of 23:49, 26 September 2012
GAL4 based yeast light-switchable promoter system
composite part of Bba_K319003, BBa_K801039, BBa_K801011 Bba_K319003, Bba_K801040, and BBa_K801011
Background and principles
This system bases on the yeast two-hybrid system which was originally created for exploring protein-protein interactions. One candidate of a potential protein-interaction pair is fused to the DNA-binding domain of a transcription factor and the other candidate to the activation domain of a transcription factor. If the proteins candidates are really physically interacting with each other, this event will starts the transcription of downstream reporter genes, e. g. LacZ or an auxotrophic marker.
Reverse yeast-two hybrid based light-switchable promoter system
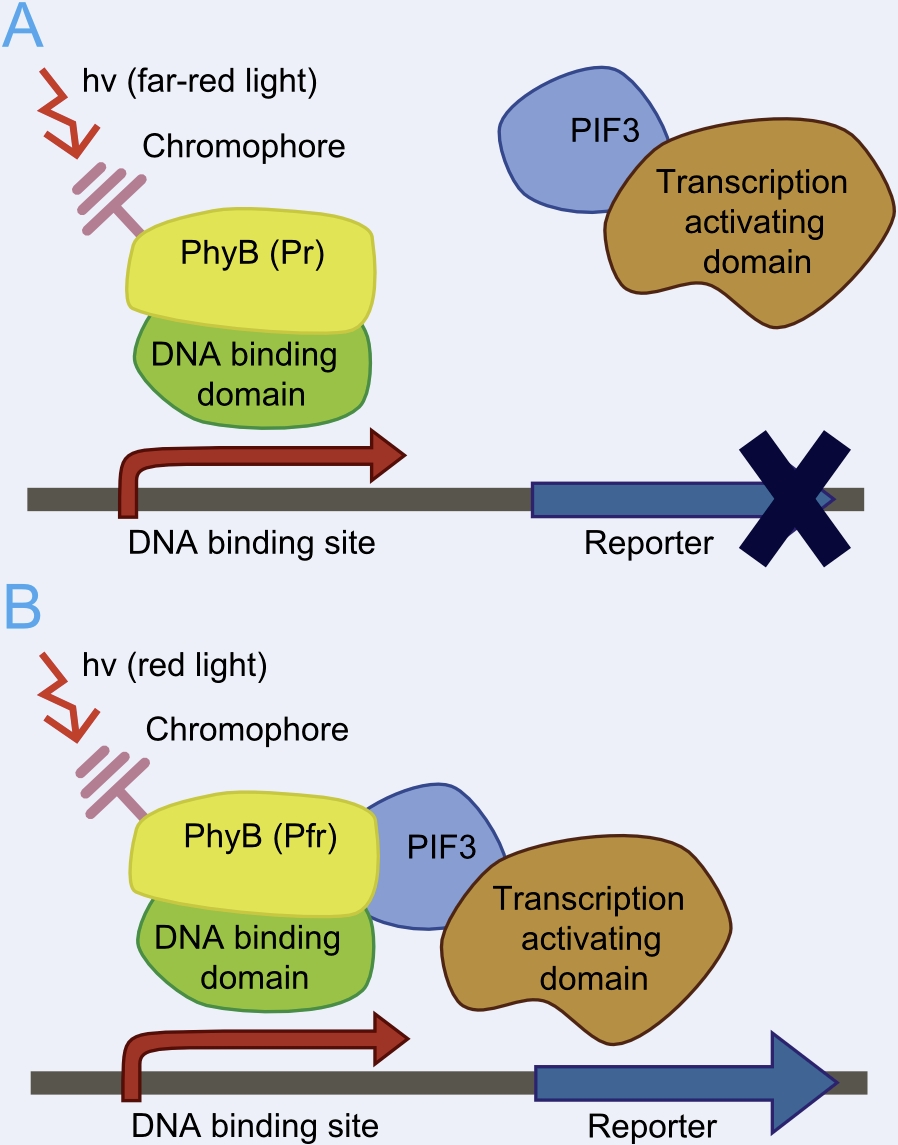
This basic principle is utilized in the yeast light-switchable promoter system. But in contrast to yeast-two hybrid, we already know the interaction partners (PhyB and PIF3). The photoconvertible binding of PhyB to PIF3 is used, to recover the physical contiguity of the DNA binding domain and the transcriptional activation domain under defined conditions (red light).
This light-inducible system contains two proteins, phytochrome B (PhyB) and phytochrome interacting factor 3 (PIF3). PhyB and PIF3 will just form a heterodimer, if PhyB is exposed to red light. Exposition under red light leads to a conformation change of PhyB to its active form (Pfr-form); the Pfr form of PhyB now can bind PIF3. PhyB comprises a light-absorbing chromophore phycocyanobilin, which gives PhyB the ability to undergo a photoconversion to the active Pfr form (red light exposition) or back to its ground-state Pr (far-red light exposition or darkness).
GAL4 based light-switchable promoter system
In our first case we create two constitutively expressed fusion proteins, the first one is PhyB fused to GAL4DBD for the DNA binding part (BBa_K801040 and the second one is PIF3 fused to GAL4AD for the transcriptional activating part (BBa_K801039). This system allows us to control spatio-temporally the expression of our genes coded on pTUM104 and driven by the GAL1 promoter (The TATA-box of pGAL1 is preceded by binding elements for GAL4). To prevent interference with the endogenous GAL4 system of yeast, we are using the Y190 S. cerevisiae strain, which has an GAL4/GAL80 deletion.
LexA based light-switchable-promoter system
For more information about the LexA based system, please see here: [parts.igem.org/wiki/index.php?title=Part:BBa_K801043 BBa_K801043]
Biosynthesis of phycocyanobilin
Phycocyanobilin undergoes a Z-E isomerization to its active form in case of red light and an E-Z isomerization to its inactive form in case of far-red light. The half-life of its active form Pfr is ~30 min, so continuous red light exposition is not nescessary. A great adventage is that light-sensitive odorant and flavorings will not be destroyed. Once phycocyanobilin is not naturally avaible in yeast one have to add the tetrapyrrole light-absorbing chromophore phycocyanobilin to the medium to get a functional light-switchable promoter system. But it also possible to bring the capability of phycocyanobilin synthesis in yeast by metabolic engineering. From heme, which is endogenous in yeast, there are only two steps of biosynthesis away from phycocyanobilin. The first step of phycocyanoblin is catalized by a heme oxygenase, the second step by a phycocyanobilin:ferredoxin oxidoreductase.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 862
Illegal BglII site found at 2795
Illegal BamHI site found at 2877
Illegal XhoI site found at 2828
Illegal XhoI site found at 2847
Illegal XhoI site found at 4982
Illegal XhoI site found at 5316 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2240
Illegal AgeI site found at 5540 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 4367
Illegal BsaI site found at 4773
Illegal BsaI site found at 5235
Illegal BsaI.rc site found at 205
Illegal BsaI.rc site found at 1986
Illegal SapI site found at 3044