Difference between revisions of "Part:BBa K777006"
| (2 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K777006 short</partinfo> | <partinfo>BBa_K777006 short</partinfo> | ||
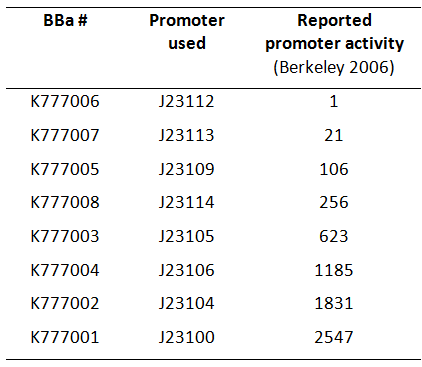
| − | + | [[Image:Table_K777001-K777008_new.png|thumb|300px|right|'''Fig. 1:''' Overview of BioBricks containing the ''Tar'' gene downstream of Anderson promoters. (Activity according to Berkeley 2006)]] | |
| − | + | ||
| − | [[Image:Table_K777001-K777008_new.png|thumb| | + | |
The transmembrane chemoreceptor Tar of <i>E. coli</i> mediates chemotaxis towards aspartate and exists as a functional homodimer. | The transmembrane chemoreceptor Tar of <i>E. coli</i> mediates chemotaxis towards aspartate and exists as a functional homodimer. | ||
| − | Here we used a set of 8 constitutive [https://parts.igem.org/Part:BBa_J23112 Anderson promoters] from the 2006 Berkeley group to test how different levels of constitutive ''Tar'' expression affect the chemotaxis of ''E. coli''. | + | Here, we used a set of 8 constitutive [https://parts.igem.org/Part:BBa_J23112 Anderson promoters] from the 2006 Berkeley group to test how different levels of constitutive ''Tar'' expression affect the chemotaxis of ''E. coli'' (Fig. 1). |
| − | <br> <br> <br> | + | <br><br> |
| − | <br> <br> <br> | + | The transcriptional activity of the constructs K777001-K777008 was quantified via quantitative real-time PCR. The results of this experiment can be found in the [https://parts.igem.org/Part:BBa_J23100:Experience#User_Reviews experience section] of the according Anderson promoters. Our data were compared to the activity of the promoters that can be found on the main page of [https://parts.igem.org/Part:BBa_J23112 Anderson promoters].<br> The respective raw dataset can be found in our Team-wiki [http://2012.igem.org/Team:Goettingen/Project/Computational_Data#Raw_Datasets here]. |
| − | <br> <br> | + | <br> <br><br> <br> <br><br> <br> <br> |
| − | <br> <br> <br> | + | |
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | This part can be used for chemotaxis assays. | |
| + | * Here is the complete sequence of this part as an [https://static.igem.org/mediawiki/2012/f/fc/Tar_receptor_under_the_control_of_constitutive_promoter.zip ApE file]. | ||
| + | <br> | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K777006 SequenceAndFeatures</partinfo> | <partinfo>BBa_K777006 SequenceAndFeatures</partinfo> | ||
| − | + | <br> | |
| + | [[Image:K777006_in_pSB1C3.png|frame|left|'''Fig. 2:''' Plasmid map of K777006 in pSB1C3]] | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 20:34, 26 September 2012
Tar receptor under the control of constitutive promoter J23112
The transmembrane chemoreceptor Tar of E. coli mediates chemotaxis towards aspartate and exists as a functional homodimer.
Here, we used a set of 8 constitutive Anderson promoters from the 2006 Berkeley group to test how different levels of constitutive Tar expression affect the chemotaxis of E. coli (Fig. 1).
The transcriptional activity of the constructs K777001-K777008 was quantified via quantitative real-time PCR. The results of this experiment can be found in the experience section of the according Anderson promoters. Our data were compared to the activity of the promoters that can be found on the main page of Anderson promoters.
The respective raw dataset can be found in our Team-wiki [http://2012.igem.org/Team:Goettingen/Project/Computational_Data#Raw_Datasets here].
Usage and Biology
This part can be used for chemotaxis assays.
- Here is the complete sequence of this part as an ApE file.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1323
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 152