Difference between revisions of "Part:BBa K392014:Experience"
(→User Reviews) |
(→User Reviews) |
||
| Line 18: | Line 18: | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| − | We ordered the C. josui CM10 (Cellulose-binding motif from C. josui Xyn10A gene) of the iGEM 2010 Osaka University to compare it to other cellulose binding domains ([https://parts.igem.org/Part:BBa_K863101 | + | We ordered the C. josui CM10 (Cellulose-binding motif from C. josui Xyn10A gene) of the iGEM 2010 Osaka University to compare it to other cellulose binding domains ([https://parts.igem.org/Part:BBa_K863101 BBa_K863101]; [https://parts.igem.org/Part:BBa_K863111 BBa_K863111]) in our project. |
In order to get the exact carbohydrate/cellulose binding domain-family of CM10 we translated the given DNA-sequence into amino acids and used the NCBI [http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&BLAST_PROGRAMS=blastp&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome Basic Local Alignment Search Tool] . The alignment the investigation showed was not a carbohydrate binding domain of any kind, but the Xylanase/Glycosyl hydrolase domain of the Xyn10A gene itself (Figure 1). | In order to get the exact carbohydrate/cellulose binding domain-family of CM10 we translated the given DNA-sequence into amino acids and used the NCBI [http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&BLAST_PROGRAMS=blastp&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome Basic Local Alignment Search Tool] . The alignment the investigation showed was not a carbohydrate binding domain of any kind, but the Xylanase/Glycosyl hydrolase domain of the Xyn10A gene itself (Figure 1). | ||
Revision as of 17:19, 20 September 2012
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K392014
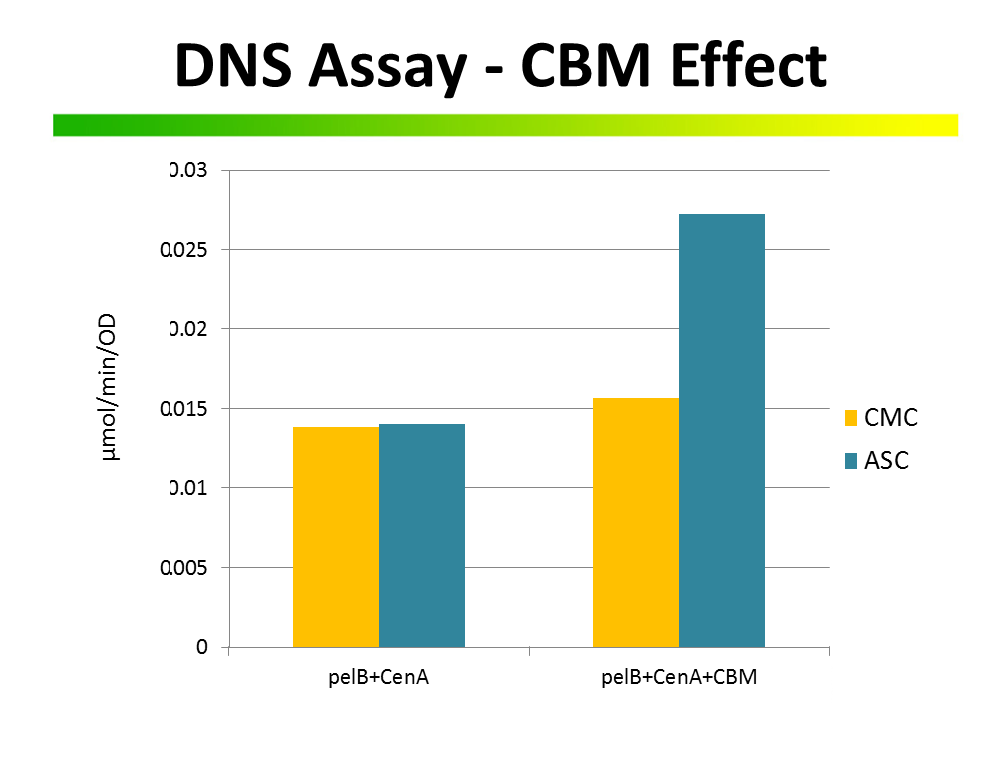
Comparison of PelB secretion tagged CenA endoglucanase with and without CM10
User Reviews
UNIQ645a60baf86ef102-partinfo-00000000-QINU
|
•
iGEM Bielefeld-Germany 2012 |
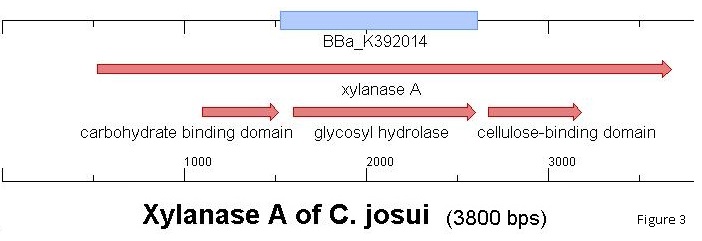
We ordered the C. josui CM10 (Cellulose-binding motif from C. josui Xyn10A gene) of the iGEM 2010 Osaka University to compare it to other cellulose binding domains (BBa_K863101; BBa_K863111) in our project. In order to get the exact carbohydrate/cellulose binding domain-family of CM10 we translated the given DNA-sequence into amino acids and used the NCBI [http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&BLAST_PROGRAMS=blastp&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome Basic Local Alignment Search Tool] . The alignment the investigation showed was not a carbohydrate binding domain of any kind, but the Xylanase/Glycosyl hydrolase domain of the Xyn10A gene itself (Figure 1). To get sure, we looked up the [http://www.ncbi.nlm.nih.gov/nuccore/AB041993 Xyn10A gene of C.josui] and searched the ORF in the same way for conserved domains. The protein search (Figure 2) showed two carbohydrate binding modules within the open reading frame and the glycosyl hydrolase domain. This result confirmed that the iGEM 2010 Osaka University had cloned the wrong domain of the xylanase-protein. The cellulose binding domain up-stream of the glycosyl hydrolase domain would have been of use for our project and is the protein-domain we would have expected by the title of this BioBrick. To match the result with the given DNA-sequence we programmed the region of every domain into a clonemanager-file of the Clostridium josui xynA gene for xylanase A and aligned it with the BBa_K392014. The alignment (Figure 3) revealed the same connection of gene and BioBrick.
|
UNIQ645a60baf86ef102-partinfo-00000002-QINU