Difference between revisions of "Part:BBa K731721"
| Line 7: | Line 7: | ||
| width=50% valign='top' style='border: 1px solid black'| | | width=50% valign='top' style='border: 1px solid black'| | ||
<partinfo>BBa_K731721 short</partinfo> | <partinfo>BBa_K731721 short</partinfo> | ||
| − | * wild type terminator from T7 | + | * wild type terminator from T7 bacteriophage. |
| − | The characterization of this terminator has been done both with T7 and E. coli RNA polymerases using [[Part:BBa_K731700|BBa_K731700]] and [[Part:BBa_K731710|BBa_K731710]] by the 2012 Trento iGEM team. | + | The characterization of this terminator has been done both with T7 and ''E. coli'' RNA polymerases using [[Part:BBa_K731700|BBa_K731700]] and [[Part:BBa_K731710|BBa_K731710]] by the 2012 Trento iGEM team. |
|} | |} | ||
Revision as of 16:48, 20 September 2012
|
T7 terminator
The characterization of this terminator has been done both with T7 and E. coli RNA polymerases using BBa_K731700 and BBa_K731710 by the 2012 Trento iGEM team. |
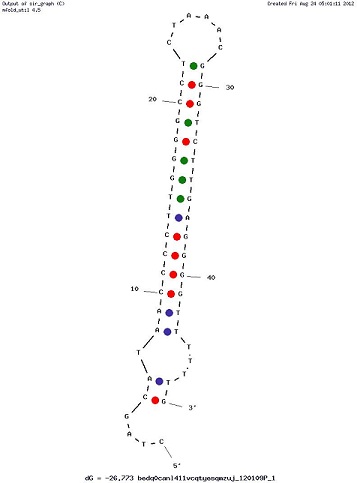
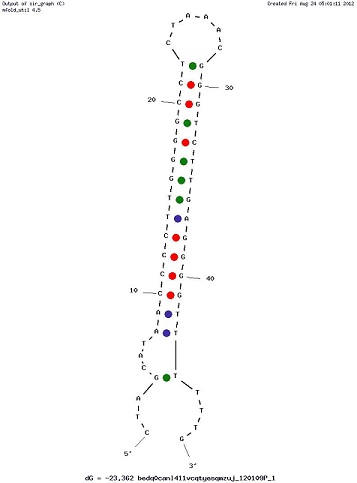
Examples of Secondary Structures
MEASUREMENTS
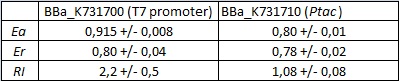
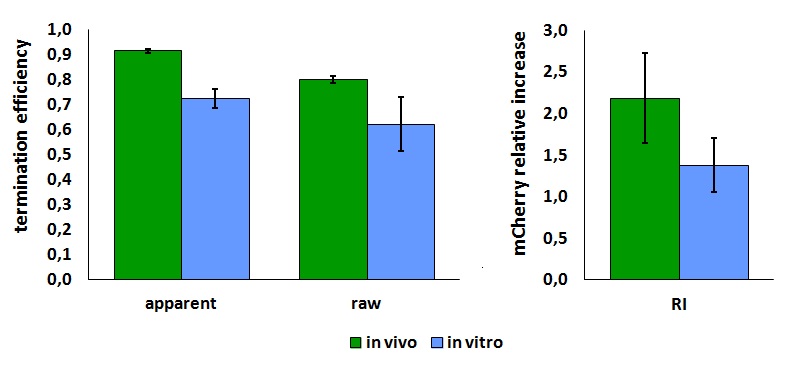
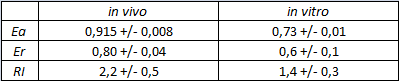
The characterization of this part was done by the Trento iGEM team 2012 using the new platforms for terminator characterization that they have built and submitted to the Registry BBa_K731700, BBa_K731710.
Its activity was analyzed both with T7(blue) and E. coli (red) RNA polymerases in vivo and with T7 RNA polymerase in vitro.
The parameters used to analyze the data are:
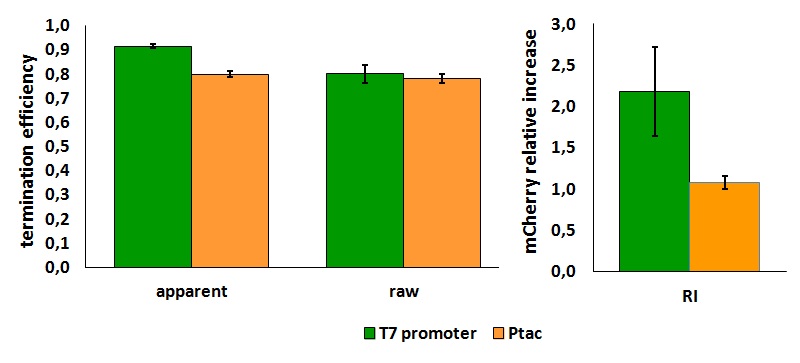
apparent termination efficiency calculated with the equation found in literature, 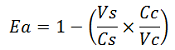
raw termination efficiency that does not consider the mCherry conribution, 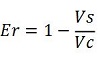
relative increase in upstream gene expression, 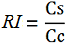
where
-Vs is the A206K Venus peak’s intensity of the construct with the terminator of interest inserted in the prefix-suffix linker
-Vc is the A206K Venus peak’s intensity of the control construct with no terminator
-Cs is the mCherry peak’s intensity of the construct with the terminator inserted
-Cc is the mCherry peak’s intensity of the control construct
In vivo measurements:
FIGURE 1. T7 bacteriophage terminator's effect on protein expression with two different RNA polymerases
The data shown in figure 1 were acquired in two different days. For each day 4 different replicates were measured at different times.
Briefly, BL21(DE3)pLysS were grown in 10 mL of LB until an OD of 0.6 was reached and induced with 0.5 mM IPTG. After 3 hours of induction, 4 separate aliquots of 1 mL were taken and sonicated 3 times for 10 seconds at intervals of 30 seconds. After sonication the samples were diluted 1:3 with PBS 1X directly into a cuvette and incubated overnight at 4°C. Fluorescence measurements were taken with a Cary Eclipse Varian fluorimeter with a window ranging from 450 nm to 700 nm using the following excitation and emission wavelengths.
In vitro measurements:
FIGURE 2. T7 bacteriophage terminator's effect on in vitro protein synthesis with the T7 RNA polymerases
Cell free measurements were done with the PurExpress kit from New England Biolabs, using 250 ng of DNA previously purified by ethanol precipitation, following the protocol suggested by the manufacturer. Measurements were done with a PTI fluorimeter using the following excitation and emission wavelengths.
More information can be found in the iGEM Trento 2012 wiki page.