Difference between revisions of "Part:BBa K640002"
Rpgguardian (Talk | contribs) |
Rpgguardian (Talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 7: | Line 7: | ||
<h3>Usage and Biology</h3> | <h3>Usage and Biology</h3> | ||
| − | + | <p>Please note that in order for proper conjugation of plasmids between <i>E. coli</i> and any other species, a proper replication origin for that species is required. For <i>Pseudomonas</i> ori1600 can be used. We submitted an optimized ori1600 part (https://parts.igem.org/wiki/index.php?title=Part:BBa_K640003) as well a composite ori1600-oriT part (https://parts.igem.org/wiki/index.php?title=Part:BBa_K640004).</p> | |
<!-- --> | <!-- --> | ||
| Line 21: | Line 21: | ||
<h3>Functionality Test</h3> | <h3>Functionality Test</h3> | ||
| − | In our first assay, we wanted to see that our BBa_K640004 (oriT-ori1600) construct was working. We incubated the co-cutures for 12 hours at 37°c prior to plating. The plates were then left to grow overnight. For further information on this part and its characterization, see our wiki page | + | In our first assay, we wanted to see that our BBa_K640004 (oriT-ori1600) construct was working. We incubated the co-cutures for 12 hours at 37°c prior to plating. The plates were then left to grow overnight. For further information on this part and its characterization, see our wiki page http://2011.igem.org/Team:Calgary/Project/Chassis/Pseudomonas. |
<h3>Results</h3> | <h3>Results</h3> | ||
Latest revision as of 20:49, 30 October 2011
oriT - Interspecies origin of transfer
oriT, or origin of transfer, is a DNA platform which allows for the transfer of plasmid DNA from one organism to another. Relaxases and nickases are responsible for cutting the DNA, host cell polymerases facilitate rolling circle replication of one of the plasmid strands in the host, and rep genes mediate transfer to the receptor organism. This oriT part is an alternate form of BBa_J01003 (optimized for transfer of plasmids between different strains of E. coli), and allows DNA transfer between E. coli and Pseudomonas.
Usage and Biology
Please note that in order for proper conjugation of plasmids between E. coli and any other species, a proper replication origin for that species is required. For Pseudomonas ori1600 can be used. We submitted an optimized ori1600 part (https://parts.igem.org/wiki/index.php?title=Part:BBa_K640003) as well a composite ori1600-oriT part (https://parts.igem.org/wiki/index.php?title=Part:BBa_K640004).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functionality Test
In our first assay, we wanted to see that our BBa_K640004 (oriT-ori1600) construct was working. We incubated the co-cutures for 12 hours at 37°c prior to plating. The plates were then left to grow overnight. For further information on this part and its characterization, see our wiki page http://2011.igem.org/Team:Calgary/Project/Chassis/Pseudomonas.
Results
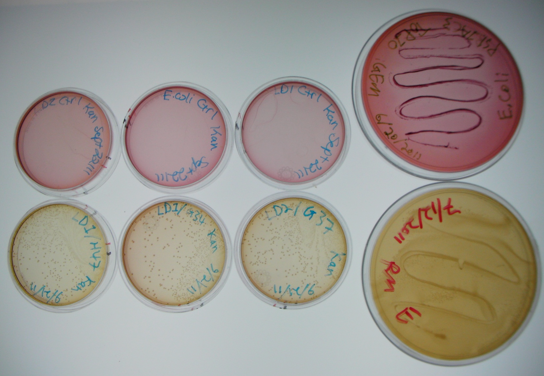
The large plates on the right show controls of Pseudomonas and E. coli on MacConkey plates indicating the normal expected colors for these organisms. The small plates on the bottom represent our construct, conjugated with two different strains of pseudomonas and plated on MacConkey agar with Kanamycin (50ug/mL). The yellow color indicates that the colonies are Pseudomonas. The small plates on the top show our two strains of Pseudomonas, as well as E. coli without kanamycin resistance plated on MacConkey agar plates with kanamycin (50 ug/mL). All of these plates are pink in color, and show no colonies. This was expected, as it shows that without a resistance marker, these organisms are not able to grow on the MacConkey kanamycin plates. This demonstrates that our part is working the way we expect it to!
