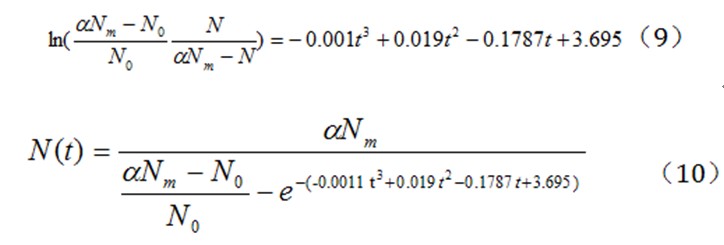Difference between revisions of "Part:BBa K658005"
| Line 123: | Line 123: | ||
| − | [[Image:XMU_China_30.jpg|left]] | + | [[Image:XMU_China_30.jpg|left|500px]] |
<html> | <html> | ||
<img src="https://static.igem.org/mediawiki/parts/4/41/XMU_China_block.jpg"> | <img src="https://static.igem.org/mediawiki/parts/4/41/XMU_China_block.jpg"> | ||
Revision as of 08:19, 4 October 2011
a bacteria population-control device with RBS0.07 driven by lacl+pL
It is designed to build a programmed bacterial death circuit, which is based on the quorum sensing system of Vibrio fischeri. iGEM-Team XMU-China has designed a series of bacteria population-control devices using RBSs of different strength in the killer protein producer. This device has the ability to maintain cell density of the bacteria population at a relatively low value compared with bacteria without this circuit and extends the steady state. RBS0.07 (BBa_B0031) was used to regulate the expression of killer protein ccdB.
Description
This device is made up of three subparts: a LuxI producer, a LuxR producer and a killer protein producer. The LuxI protein synthesizes a small, diffusible acyl-homoserinelactone (AHL) signaling molecule. The AHL accumulates as the cell density increases. At sufficiently high concentrations, it binds the LuxR, which induces the expression of the killer gene ccdB under the control of a promoter lux pR. Sufficiently high levels of CcdB which is a bacterial toxin that targets DNA gyrase cause cell death. Low cell density doesn’t have the ability to produce sufficient LuxR/AHL complex to activate the promoter lux pR. The programmed death circuit ends and the cell density increases. When the cells reach a certain concentration, the death circuit is restarted. Back and forth, the programmed death is achieved in the dynamic process of growth and death. In that way, the bacteria population is programmed to maintain one certain cell density.
Performance
In order to test the performance of the bacteria population-control device iccdB0.07, this device was first cloned into plasmid pSB1A2, followed by transformation into E.coli strain BL21.
The cell growth curves in Figure 2 showed the bacteria population-control device successfully maintained the cell density at a lower value at the steady state compared with BL21’s cells without this circuit. Besides, circuit-regulated cell growth (black dot) has a relatively longer steady state than cells without this circuit (red dot).
For iccdB0.07 regulated cell growth, the viable cell density first start to decline around 8h and then reached a steady state after two minor oscillations. Compared with bacterium without this circuit, the iccdB0.07 regulated bacterium has a lower media-consuming rate due to its lower cell density.
The iccdB0.07 regulated cell growth might be explained by “ON-OFF” mechanism based on the quorum sensing system.

iGEM Team XMU-China has also tested the performance of three other bacteria population-control device iccdB1.0 (BBa_K658001), iccdB0.6 (BBa_K658003) and iccdB0.3 (BBa_K658004). Figure 3, figure 4 and table 1 illustrate that by using RBS of different strength in the population-control device, we were able to control the steady-state cell density of a bacteria population at different levels. And a population-control device with RBS of high strength results in a low steady-state cell density. It might be explained by the mechanism that for circuit-regulated growth, the cell death rate is regarded proportional to the intracellular concentration of the killer protein. The expression of the killer gene is regulated by the strength of its upstream RBS. Therefore, a RBS with higher strength promises more killer protein in vivo, which leads to a higher death rate of the bacteria population.
Model
Different RBS sequences leads to different levels of expression of the killer protein CcdB which is directly linked to the effects of our programmed cell-death circuit. So we constructed a series of circuits with different RBS sequences so as to detect how RBS of different efficiency can affect the viable cell density at steady state. We build a model to search for a theory to predict growth rule of bacteria with the programmed cell-death circuit. Logistic kinetics is widely used in describing the growth rule of bacteria. Based on that, bacteria without the cell-death circuit follow the equation (1) listed below:

When viable cell density reaches the steady state, the equation (2) can be applied:

k is the growth rate constant 1/h,β is the death coefficient. N_m is the carrying capacity in the Limited medium without the cell-death circuit.
In our experiment,we find that the viable cell density at steady-state Ne is lower than the Nm with the cell-death circuit(Figuer 5).
Assume that:

With time limited, we only conducted the experiment on the growth curve of BL21 with the population-control device with RBS0.07 and the viable cell density at steady-state with the population-control device with RBS0.3, RBS0.6 and RBS1.0.
Computed from our experimental data, we can get the data listed in Table 2 and Figure 5:

The following model is built based on the data of the population-control device with RBS0.07. The value α=0.692 can be applied in the following calculation.
It is assumed that both k(t) and β(t) are the functions of time.

Equation (6) is derived from equation (1)(3)(4)(5):

By substituting the value of N corresponding to different t, we can get the value of

Then we can draw the plot in the graph in which

and t represent vertical and horizontal coordinates. A fitting curve can be obtained using the excel. Thus, we can get the value of k, a and b.

Figure 6 We can get the following equations (9) and (10) from the Figure 6

Method
1.Isolation of Plasmid
Using the Procedure for GenEluteTM Plasmid Miniprep Kit
- Collect 1-5 mL bacterium fluid in 1.5 mL centrifuge tube,and centrifuge fluid at 12000 r/min
- Resuspend cells. Discard the supernatant and completely resuspend the bacterial pellet with 250 µl of the Resuspension Solution
- Lyse cells. Lyse the resuspended cells by adding 250 µl of the Lysis Solution
- Neutralize. Precipitate the cell debris by adding 350 µl of the Neutralization/Binding Solution, and centrifuge fluid at 12000r/min
- Load cleared lysate. Transfer the supernatant from step 4 to the spin column. Centrifuge at 12000r/min for 1 minute, and then discard the filtrate
- Optional wash. Add 500 µl of the Optional Wash Solution to the column. Centrifuge at 12000 r/min for 1 minute. Discard the filtrate
- Wash column. Add 500 µl of the diluted Wash Solution to the column. Centrifuge at 12000r/min for 1 minute.
- Elute DNA. Transfer the column to a new collection tube. Add 50~100 µl of Eluent Solution to the column. Centrifuge
at 12000 r/min for 1 minute. The DNA is now present in the filtrate and is ready for immediate use or storage at -20℃
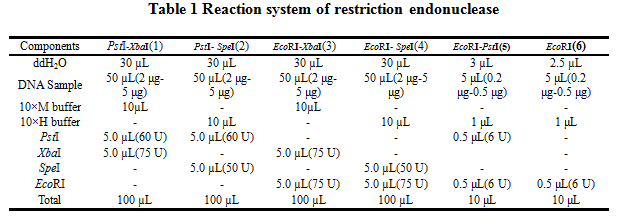
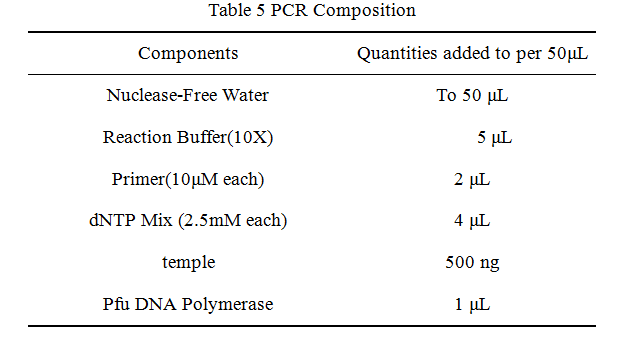
2.Reaction system of restriction endonuclease

System1、2、3 and 4 are used for Standard BioBrick Assembly .System 5 and 6 are used for Restriction analysis. Digestion of sample: at least 500 ng DNA / 10 µL volume. Digest for 4 h at 37 °C, afterwards inactivated by adding 10x loading buffer and standing for 10 min at room temperature.
3. Standard BioBrick Assembly
- Digestion of insert: 2 μg~5 μg DNA / 100 µL volume, 10x H buffer, EcoRI, SpeI. Digestion and inactivation. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel, try to avoid staining or exposure to ultraviolet light of the insert.
- Digestion of vector: 2 μg~5 μg DNA / 100 µL volume, 10 x M buffer, EcoRI, XbaI. Digestion and inactivation. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel, try to avoid staining or exposure to ultraviolet light of the insert.
4. Suffix Insertion
- Digestion of insert: 2 μg~5 μg DNA / 100 µL volume, 10x M buffer, XbaI, PstI. Digestion and inactivation. Clean up the insert.
- Digestion of vector : 2 μg~5 μg DNA / 100 µL volume, 10x H buffer, SpeI, PstI. Digestion and inactivation. Clean up the vector.
5. Ligation
- After digestion and clean-up, the next step is ligation. Overnight ligation at 16°C. Table 2 is the system of ligation.
6. Transformation
- Preparation of competent E.coli cells
- Add 10 µL plasmid to 100 µl competent cells in centrifuge tube
- Store tube on ice for 20-30 minutes
- Water bath for 90s at 42℃
- Put the tube on ice for 1-2min
- Add 790 µL LB,and cultivation for 1h at 37 ℃,then plate on selective LB-Medium.
7. Restriction analysis
- Pick one colony with a sterile tip and cultivation in 20ml LB for overnight at 37 ℃
- Isolation of Plasmid
- Digest BioBrick,the system of Restriction analysis refer to table1
- Gel electrophoresis:add 2 µL loading buffer to digestion mixture. An agarose concentration is 1 %.
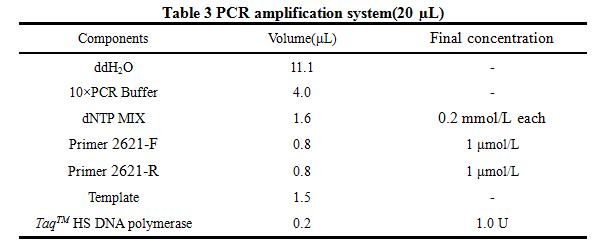
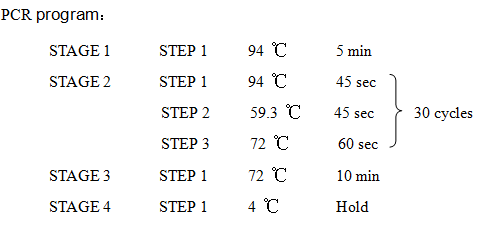
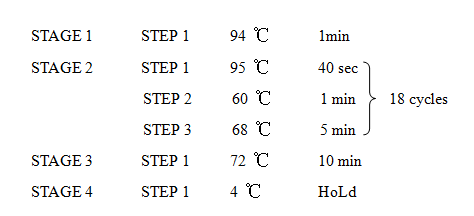
8. PCR -RBS1.0-luxR-TT-lux pR-
Design Primers using the primer 5
Primer:
2621-F:5'-GCTCTAGAGAAAGAGGAGAAATACT-3'
2621-R:5'-AAAACTGCAGCGGCCGCTACTA-3'

9. Cell growth
- 100µL suspension was inoculated from a Glycerin tube into 20ml fresh LB and incubated overnight at 37℃ and 250 r.p.m.
- 100µL suspension was inoculated again from step1 into 50ml fresh LB and incubated at 37℃ and 250r.p.m
- IPTG was added when A600≈0.6-0.8
- 1 ml suspension was taken on every sample taken time. 3 samples were taken in each time.
- Diluted each sample to 10-6(Sometimes 10-5),and then plate on selective LB-Medium.
- After 12h, count the number of CFU on the plate on different time point and then draw the cell growth curve.

10. Determining fluorescence intensity
- Add IPTG when A600 0.6~0.8.
- Cool the culture 10 minutes on ice.
- Centrifuge at 6000rpm. Wash it with pre-cooled PBS buffer.
- Use fluorescence spectrophotometer tomeasure the fluorescenceof GFP:
- Before measuring, dilute the bacteria with PBS buffer so that it can be within the measuring #range. Set excitation wavelength 491 nm, emission wavelength 511 nm.
- Transfer the measured bacteria in a new centrifuge tube and measure the OD of the bacteria.
11. Site Directed Mutagenesis

•Digest the template plasmid by adding 1 µL of DpnI and incubate for 1-2 h
•Transform 10 µL of t PCR product into competent E. coli cells
•Screen the transformants using restiction digest and sequencing
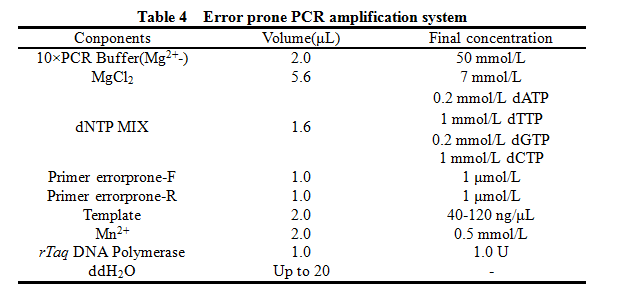
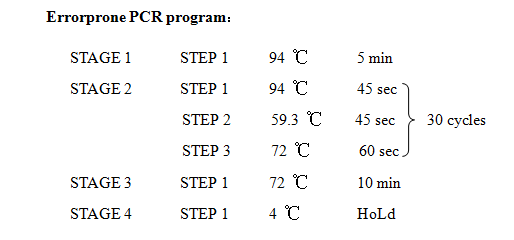
12. Error prone PCR

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 718
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 2346
Illegal BsaI.rc site found at 1874