Difference between revisions of "Part:BBa K678002"
(→Usage and Biology) |
|||
| (9 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K678002 short</partinfo> | <partinfo>BBa_K678002 short</partinfo> | ||
| − | + | This is a device intended for expression of green fluorescent protein (GFP) in the peroxisomes of mammalian cells. | |
| + | The expression of GFP is under the control of the strong constitutive cytomegalovirus (CMV) promoter. The targeting sequence S-K-L often referred to as peroxisomal targeting sequence 1 (PTS1) is directly fused to the C-terminal of GFP, this sequence should ensure the localization of GFP to the peroxisomes of the cell. | ||
| + | |||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
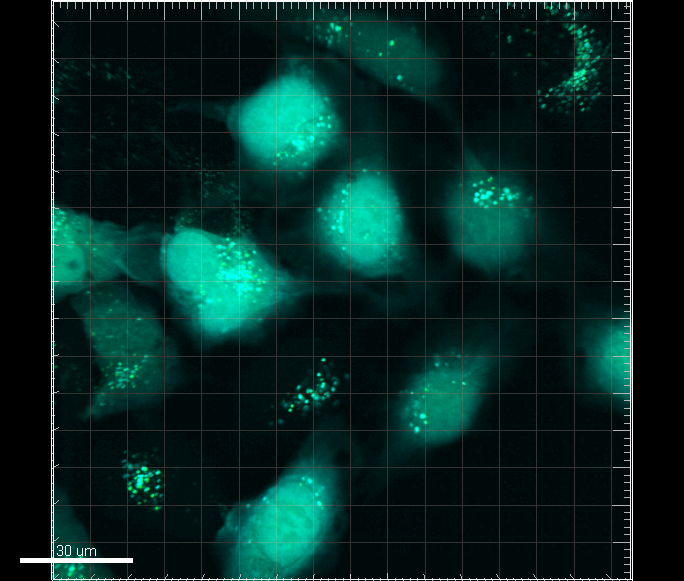
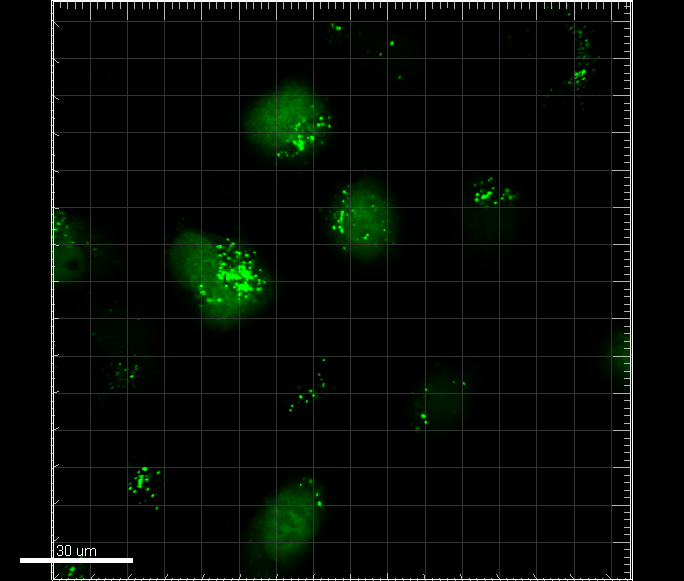
| + | We used this device to target enhanced GFP to the peroxisomes of the mammalian cell line U-2 OS (Figure 1). Although the microscopy shows GFP distributed in points which most likely are the peroxisomes of the cells, further analysis are necessary to confirm that GFP is in fact is targeted to the peroxisomes. We furthermore transiently transfected U-2 OS with both the device contained on plasmid pJEJAM2 and with pJEJAM5 expressing cyan fluorescent protein (CFP) (Figure 2). | ||
| + | |||
| + | |||
| + | [[Image:Gfppts.png|left|400px|thumb|<b>DTU-Denmark-2 2011</b> Figure 1: U-2 OS cells transiently transfected with [https://parts.igem.org/wiki/index.php?title=Part:BBa_K678050 pJEJAM2] expressing GFP localized to the peroxisomes. As can be seen on the picture this vector most likely localizes GFP to the peroxisomes of the cells. The white bar has a length of 30μm.]] | ||
| + | |||
| + | [[Image:Gfpptscfp.png|right|400px|thumb|<b>DTU-Denmark-2 2011</b> Figure 2: U-2 OS cells transiently transfected with [https://parts.igem.org/wiki/index.php?title=Part:BBa_K678050 pJEJAM2] and [https://parts.igem.org/wiki/index.php?title=Part:BBa_K678053 pJEJAM5] expressing GFP localized to the peroxisomes and CFP, respectively. The white bar has a length of 30μm.]] | ||
| + | |||
| + | <br clear=all> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 10:24, 27 September 2011
eGFP-PTS1 for mammalian cells
This is a device intended for expression of green fluorescent protein (GFP) in the peroxisomes of mammalian cells. The expression of GFP is under the control of the strong constitutive cytomegalovirus (CMV) promoter. The targeting sequence S-K-L often referred to as peroxisomal targeting sequence 1 (PTS1) is directly fused to the C-terminal of GFP, this sequence should ensure the localization of GFP to the peroxisomes of the cell.
Usage and Biology
We used this device to target enhanced GFP to the peroxisomes of the mammalian cell line U-2 OS (Figure 1). Although the microscopy shows GFP distributed in points which most likely are the peroxisomes of the cells, further analysis are necessary to confirm that GFP is in fact is targeted to the peroxisomes. We furthermore transiently transfected U-2 OS with both the device contained on plasmid pJEJAM2 and with pJEJAM5 expressing cyan fluorescent protein (CFP) (Figure 2).
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 2233
Illegal SpeI site found at 49
Illegal PstI site found at 2323 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 2233
Illegal SpeI site found at 49
Illegal PstI site found at 2323 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 2233
- 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 2233
Illegal SpeI site found at 49
Illegal PstI site found at 2323 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 2233
Illegal SpeI site found at 49
Illegal PstI site found at 2323
Illegal NgoMIV site found at 3088 - 1000COMPATIBLE WITH RFC[1000]