Difference between revisions of "Part:BBa K510022"
| (5 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
| − | + | '''Single-target transposition to the bacterial genomes''' is dependent on the presence of a conserved sequence, encompasing the 3' end of the glmS gene, encoding glucosamine synthetase, and part the intergenic region immediately downstream, designated the '''attTn7 site''' (from attachment site). Our [http://2011.igem.org/Team:UPO-Sevilla/Foundational_Advances/MiniTn7/Bioinformatics/attTn7_Insertion_Site Bioinformatic analysis] reveal that this region is highly conserved, and Tn7 transposition has been demonstrated in over 20 bacterial species ([http://www.ncbi.nlm.nih.gov/pubmed/8556868 Craig, 1996]). However, the lack of a suitable target may be the limiting factor in some of the organisms in which Tn7 transposition does not occur, including a variety of bacteria, but also archea and eukaryotes. The possibility of inserting a functional attTn7 in the genomes of these organisms may enable site-specific Tn7 transposition for at least some of them, thus expanding the host range of this transposon and its derived tools, such as the [http://2011.igem.org/Team:UPO-Sevilla/Foundational_Advances/MiniTn7/Overview miniTn7 BioBrick toolkit] ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K510000 BBa_K510000], [https://parts.igem.org/wiki/index.php?title=Part:BBa_K510012 BBa_K510012]). | |
| − | + | ||
| + | [[Image:UPOSevilla-attTn7-glmS.jpg|700px|center]] | ||
| + | |||
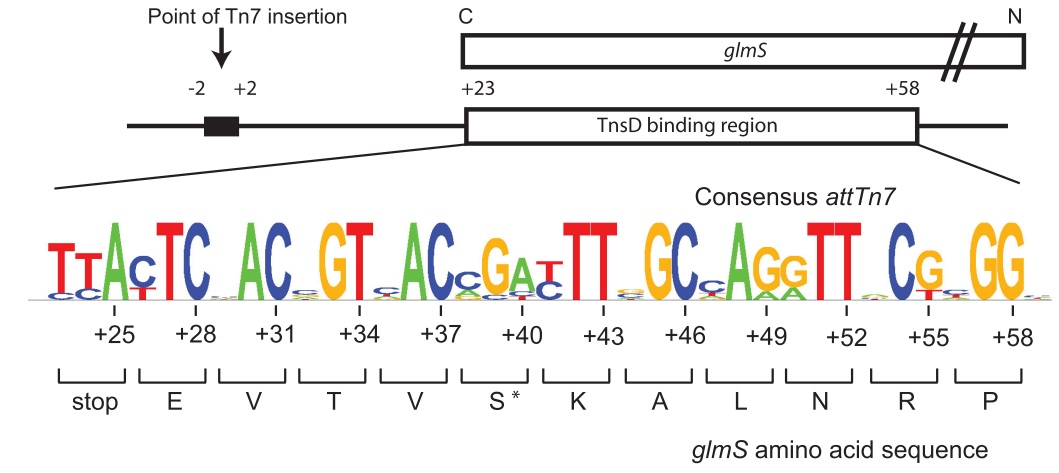
| + | '''Figure. The organization of attTn7''' (from [http://www.mobilednajournal.com/content/1/1/18 Rupak et al, 2010]). A schematic of the attTn7 at the C-terminus of the glmS gene with the TnsD binding site is shown. The sequence of the E. coli GlmS protein, and the consensus attTn7 sequence were derived as described in the text (note that the least conserved nucleotides correspond to the third position of each codon). | ||
| + | |||
| + | More information about this plasmid from the UPO-Sevilla 2011 team in the ''Experience section''. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 22:41, 24 September 2011
attTn7
Single-target transposition to the bacterial genomes is dependent on the presence of a conserved sequence, encompasing the 3' end of the glmS gene, encoding glucosamine synthetase, and part the intergenic region immediately downstream, designated the attTn7 site (from attachment site). Our [http://2011.igem.org/Team:UPO-Sevilla/Foundational_Advances/MiniTn7/Bioinformatics/attTn7_Insertion_Site Bioinformatic analysis] reveal that this region is highly conserved, and Tn7 transposition has been demonstrated in over 20 bacterial species ([http://www.ncbi.nlm.nih.gov/pubmed/8556868 Craig, 1996]). However, the lack of a suitable target may be the limiting factor in some of the organisms in which Tn7 transposition does not occur, including a variety of bacteria, but also archea and eukaryotes. The possibility of inserting a functional attTn7 in the genomes of these organisms may enable site-specific Tn7 transposition for at least some of them, thus expanding the host range of this transposon and its derived tools, such as the [http://2011.igem.org/Team:UPO-Sevilla/Foundational_Advances/MiniTn7/Overview miniTn7 BioBrick toolkit] (BBa_K510000, BBa_K510012).
Figure. The organization of attTn7 (from [http://www.mobilednajournal.com/content/1/1/18 Rupak et al, 2010]). A schematic of the attTn7 at the C-terminus of the glmS gene with the TnsD binding site is shown. The sequence of the E. coli GlmS protein, and the consensus attTn7 sequence were derived as described in the text (note that the least conserved nucleotides correspond to the third position of each codon).
More information about this plasmid from the UPO-Sevilla 2011 team in the Experience section.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]