Difference between revisions of "Part:BBa K525222"
(→Identification and localisation) |
(→Identification and localisation) |
||
| Line 188: | Line 188: | ||
| − | As expected, no sequence coverage was found in the periplasmatic fraction, due to absent tat sequence located at the amino-terminus. Little fluorescence was also found in the fraction of supernatant of the media, indicating that the protein can not be transported to the | + | As expected, no sequence coverage was found in the periplasmatic fraction, due to absent tat sequence located at the amino-terminus. Little fluorescence was also found in the fraction of supernatant of the media, indicating that the protein can not be transported to the periplasm and thus secretion into the medium does not take place. The denaturation fraction and the Triton X-100 fraction show no or very few sequence coverage, however the lysis fraction shows significant higher sequence coverage. Both results indicate, that the fusion protein solely present in the cytoplasm and thus only identified in the lysis fraction. Fig. 5 and FIg. 6 show that the protein can be found mainly in the lysis fraction, but in smaller amounts ind the periplasmatic and the media fraction, which can be explained due to the abscence of the lipid anchor. The anchor normally binds to the cell membrane, so no protein is found in other than the lysis fraction. |
To obtain more specific informations about the location of the S-layer fusion protein, after comparison with same treated fraction of ''E. coli'' KRX all gel bands in a defined size area were cut out of the gel and analysed with MALDI-TOF. Results are shown in fig. 6. | To obtain more specific informations about the location of the S-layer fusion protein, after comparison with same treated fraction of ''E. coli'' KRX all gel bands in a defined size area were cut out of the gel and analysed with MALDI-TOF. Results are shown in fig. 6. | ||
Revision as of 01:15, 22 September 2011
S-layer cspB from Corynebacterium halotolerans
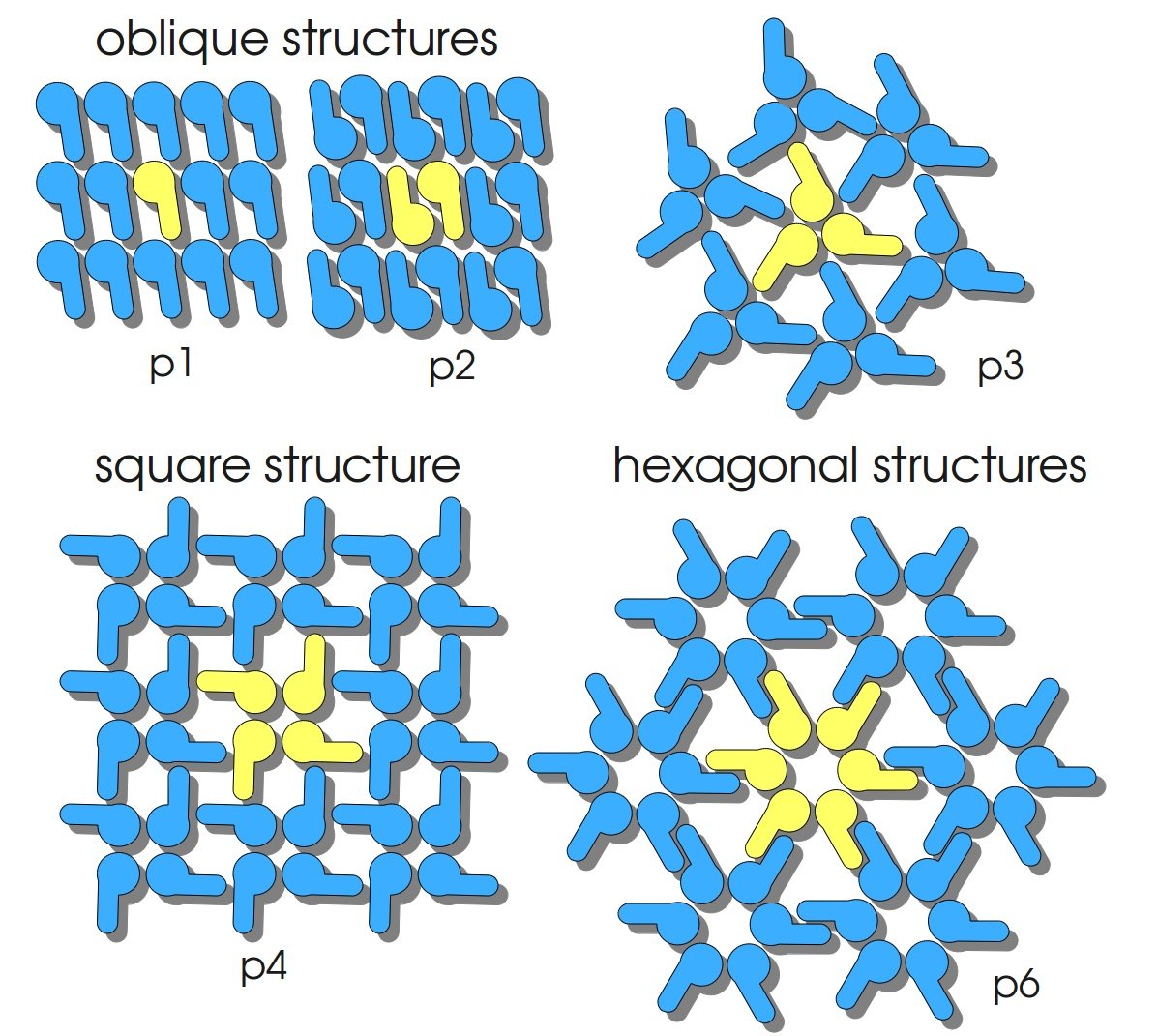
S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self assembly-domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to eachother (determined by the S-layer geometry) it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr et al., 2007]).
Usage and Biology
S-layer proteins can be used as scaffold for nanobiotechnological applications and devices by e.g. fusing the S-layer's self-assembly domain to other functional protein domains. It is possible to coat surfaces and liposomes with S-layers. A big advantage of S-layers: after expressing in E. coli and purification, the nanobiotechnological system is cell-free. This enhances the biological security of a device.
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression (E. coli) | Localisation | Cell membrane |
| Compatibility | E. coli KRX | |
| Induction of expression | L-rhamnose for induction of T7 polymerase | |
| Specific growth rate (un-/induced) | 0.245 h-1 / 0.109 h-1 | |
| Doubling time (un-/induced) | 2.83 h / 6.33 h |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1103
Illegal XhoI site found at 559 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 226
Illegal NgoMIV site found at 1315
Illegal AgeI site found at 217
Illegal AgeI site found at 458
Illegal AgeI site found at 505 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 907
Illegal BsaI.rc site found at 214
Illegal BsaI.rc site found at 592
Illegal BsaI.rc site found at 994
Expression in E. coli
The CspB gen was fused with a monomeric RFP (BBa_E1010) using [http://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly] for characterization.
The mRFP|CspB fusion protein was overexpressed in E. coli KRX after induction of T7 polymerase by supplementation of 0,1 % L-rhamnose using the [http://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#Expression_of_S-layer_genes_in_E._coli autinduction protocol] from promega.
Identification and localisation
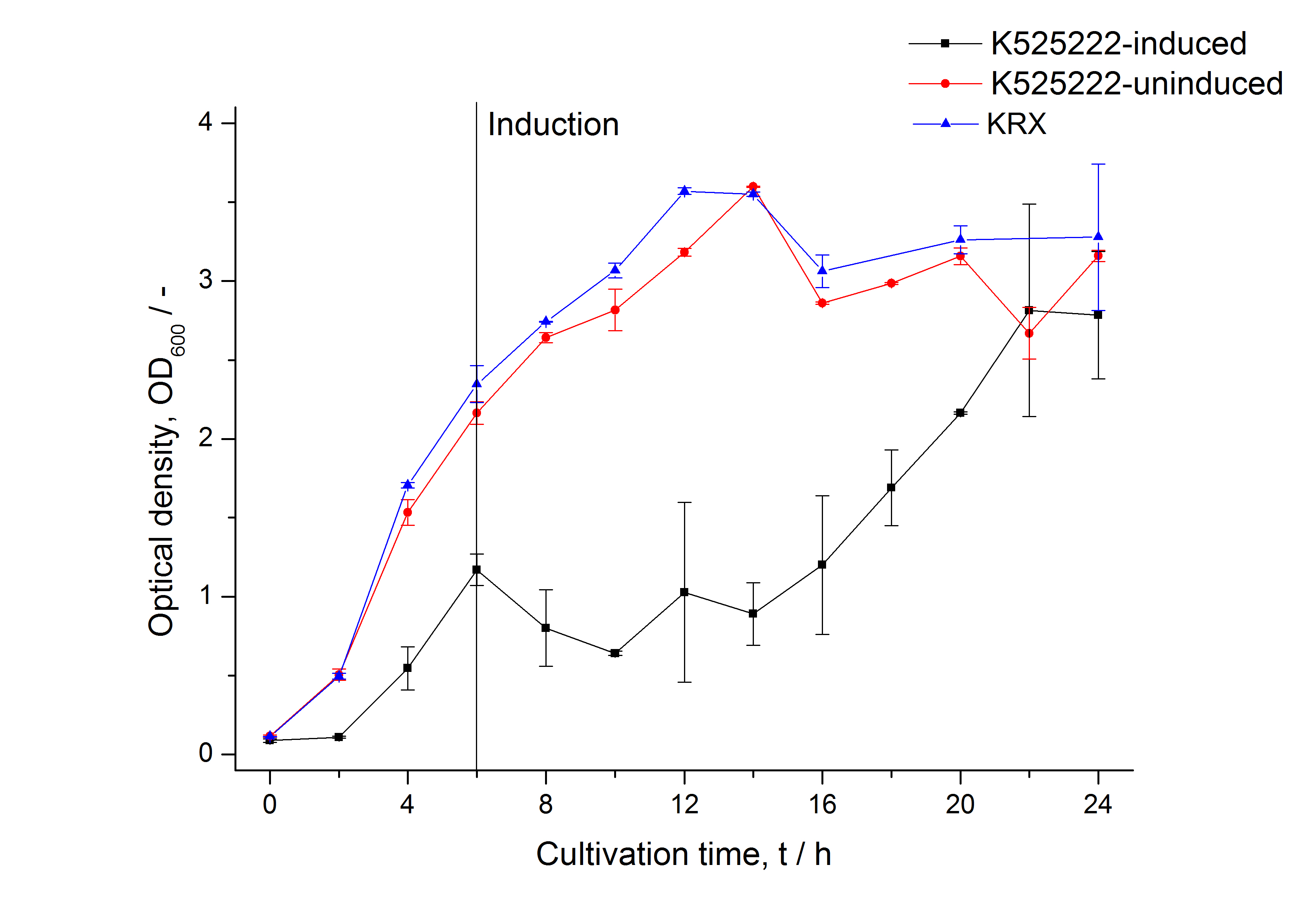
After a cultivation time of 18 h the mRFP|CspB fusion protein has to be localized in E. coli KRX. Therefor a part of the produced biomass was mechanically disrupted and the resulting lysate was wahed with ddH2O. Then the lysate was treted with ionic, nonionic and zwitterionic detergents to release the CspB|mRFP out of the membranes, if it intigrates. From the other part of the cells the periplasm was detached by using a osmotic shock.
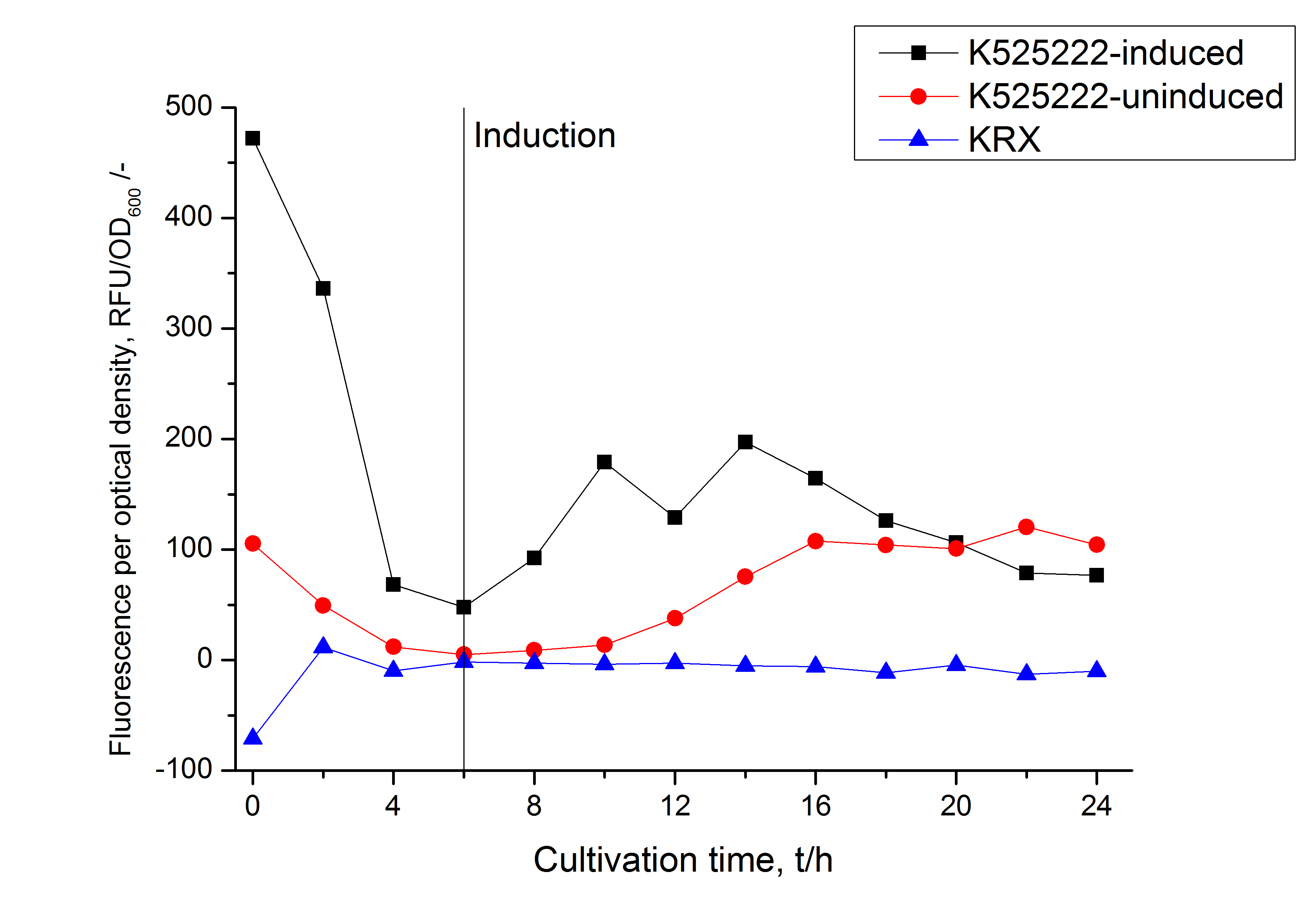
The fluorescence in all cultivation fractions plus the fluorescence in the lysis und wash fraction shows that the fusion protein is water soluble and sediment not during centrifugation. Together with the absence of flourescence in the detergent fractions verifies that the fusion protein is not integrated into the cell membrane (fig. 3) and is not present as inclusion body.
In comparison with the mRFP fusion protein of ???, wich has a lipid anchor, a minor relative fluorescence per OD600 in all cultivation and detergent fractions was detected (fig. 3). Together with the decreasing RFU/OD600 after 14 h of cultivation (fig. 2) indicates this rusult a postive effect of the lipid anchor on the protein stability.
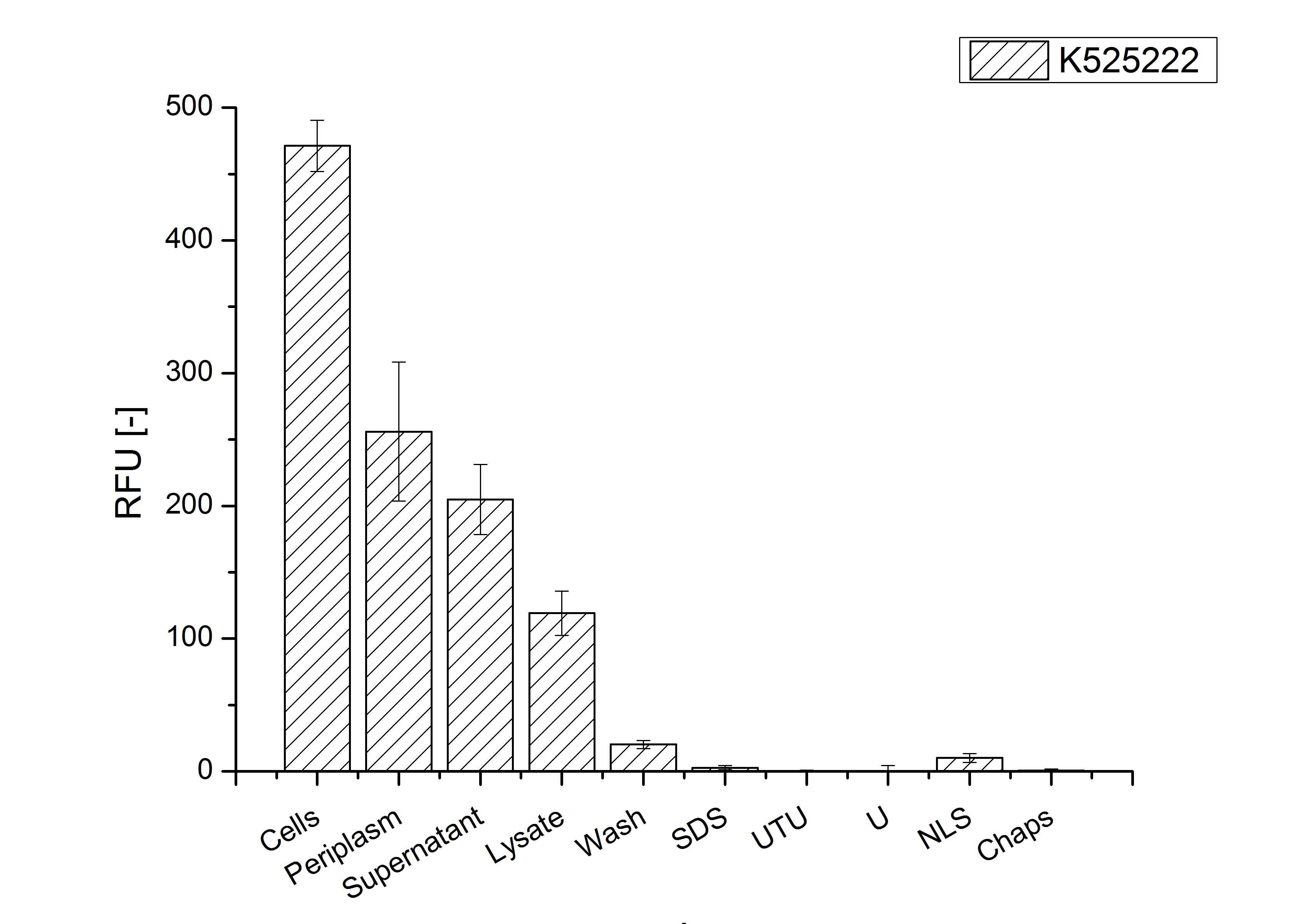
MALDI TOF analysis was used to identify the location of the fusion protein in different fractions. Fractions of medium supernatant after cultivation, periplasmatic isolation, cell lysis and following denaturation in 6 M urea were loaded onto a SDS_PAGE. After denaturation with 6 M urea the remaining pellet (after centrifugation 15,000 g for 30 min) was washed with 2 % (v/v) Triton X-100, 2 % SDS (w/v). This fraction was also loaded onto the SDS-PAGE and fragments of the gel were measured with MALDI TOF.
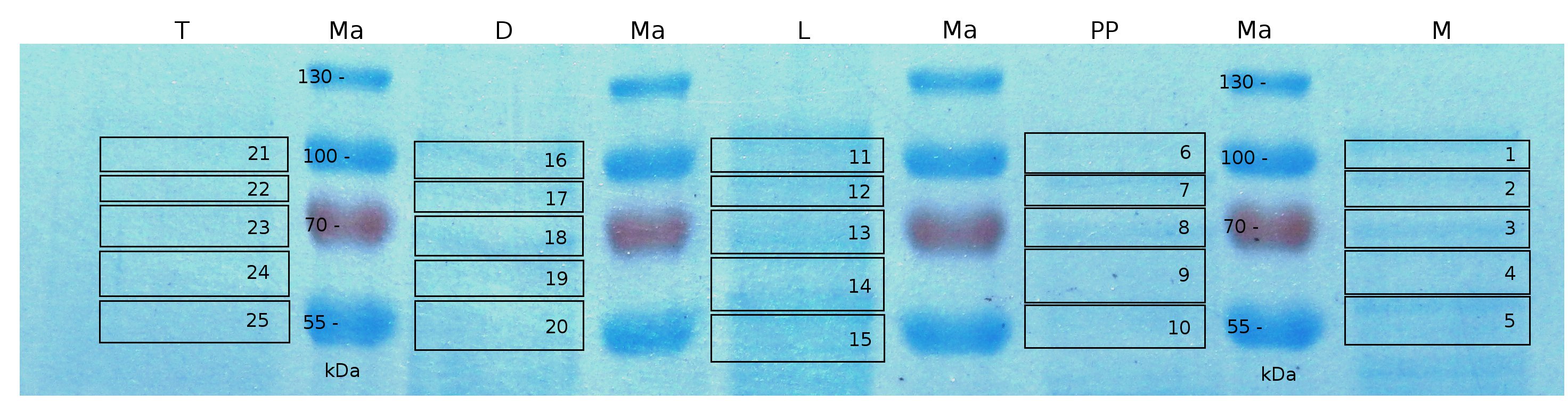
The following table shows the sequence coverage (in %) of our measurable gel samples with the amino acid sequence of fusion protein CspB/mRFP (BBa_E1010).
| number of gel sample | sequence coverage (%) |
|---|---|
| 1 | 0.0 |
| 2 | 0.0 |
| 3 | 0.0 |
| 4 | 0.0 |
| 5 | 0.0 |
| 6 | 0.0 |
| 7 | 0.0 |
| 8 | 0.0 |
| 9 | 0.0 |
| 10 | 0.0 |
| 11 | 0.0 |
| 12 | 14.6 |
| 13 | 9.3 |
| 14 | 6.3 |
| 15 | 0.0 |
| 16 | 0.0 |
| 17 | 0.0 |
| 18 | 0.0 |
| 19 | 0.0 |
| 20 | 1.0 |
| 21 | 0.0 |
| 22 | 0.0 |
| 23 | 0.0 |
| 24 | 0.0 |
| 25 | 0.0 |
Fig. 5 shows these data. The gel samples were arranged after estimated molecular mass cut out from the gel.
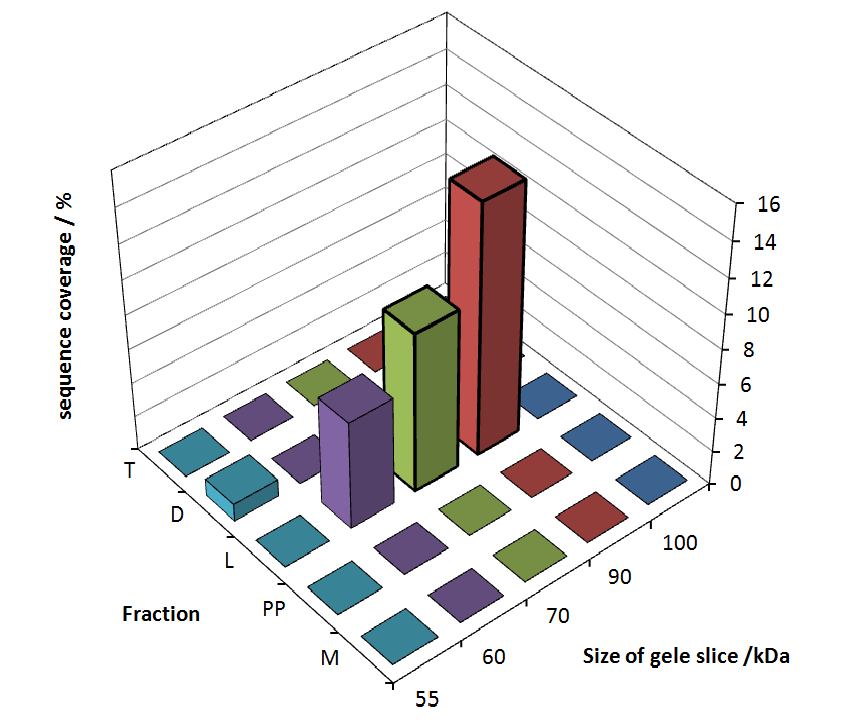
As expected, no sequence coverage was found in the periplasmatic fraction, due to absent tat sequence located at the amino-terminus. Little fluorescence was also found in the fraction of supernatant of the media, indicating that the protein can not be transported to the periplasm and thus secretion into the medium does not take place. The denaturation fraction and the Triton X-100 fraction show no or very few sequence coverage, however the lysis fraction shows significant higher sequence coverage. Both results indicate, that the fusion protein solely present in the cytoplasm and thus only identified in the lysis fraction. Fig. 5 and FIg. 6 show that the protein can be found mainly in the lysis fraction, but in smaller amounts ind the periplasmatic and the media fraction, which can be explained due to the abscence of the lipid anchor. The anchor normally binds to the cell membrane, so no protein is found in other than the lysis fraction.
To obtain more specific informations about the location of the S-layer fusion protein, after comparison with same treated fraction of E. coli KRX all gel bands in a defined size area were cut out of the gel and analysed with MALDI-TOF. Results are shown in fig. 6.
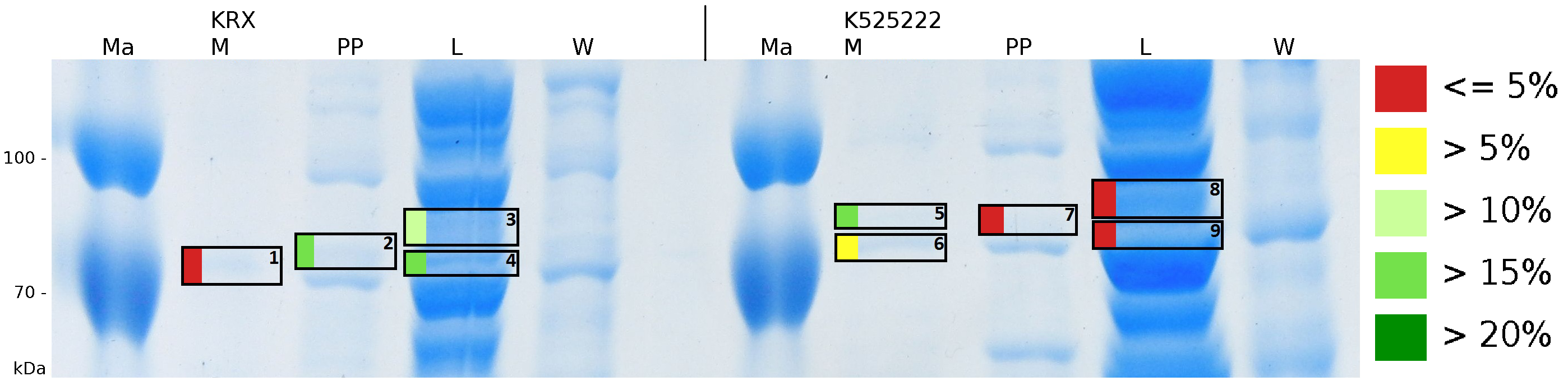
MALDI TOF measurement shows sequence coverage in the supernatant fraction of the medium, the periplasmatic fraction and the lysis fraction, indicating that the fusion protein of K525222 and BBa_E1010 without TAT-sequence and lipid anchor mostly found in the cytoplasm. The periplasmatic isolation does destroy few complete cells, therefore in the periplasmatic fraction sequence coverage is found. The gel lanes in the periplasmatic fraction are clearly less intense, the protein concentration is very low compared to the lysis fraction. Sequence coverage in the media fraction shows, that the protein is probably secreted into the media.