Difference between revisions of "Part:BBa K638202"
| (5 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
Arabinose inducible Poly-His Reflectin A1 generator produces a poly-histidine (his-tagged) form of Reflectin A1([[Part:BBa_K638001 | BBa_K638001]]). | Arabinose inducible Poly-His Reflectin A1 generator produces a poly-histidine (his-tagged) form of Reflectin A1([[Part:BBa_K638001 | BBa_K638001]]). | ||
| − | |||
| − | + | Bacteria expressing this construct will produce Reflectin A1 with a poly-histidine tag at the N-terminus (upon induction with arabinose) in order to enable purification with a his-affinity column used in conjunction with nickel or cobalt resins. | |
| − | + | ||
| + | The Reflectin A1 gene is under the control of the pBAD promoter [[Part:BBa_I0500 | BBa_I0500]], which is tightly controlled by two factors: | ||
| + | *'''L-arabinose''' monosaccharide taken up by the cell from the medium, which acts as an ''inducer''. | ||
| + | *'''AraC''' protein included in the I0500 biobrick, which acts an a ''repressor''. | ||
| + | Therefore, the araC-pBAD system offers regulatable control of gene expression in the presence of the inducer and highly repressed in the absence of the inducer. | ||
| + | Read more about [http://2011.igem.org/Team:Cambridge/Experiments/Low_Level_Expression the pBAD and arabinose system in E.coli]. | ||
| + | |||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K638202 SequenceAndFeatures</partinfo> | <partinfo>BBa_K638202 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | ===Usage and Biology=== | ||
| + | Best used in an ''E. coli'' chassis such as strain [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783], with constitutive expression of an Arabinose transporter. | ||
| + | See our [[Part:BBa_I0500:Experience | experience of pBAD]] for some issues with tuning expression levels. | ||
| + | |||
| + | See [http://2011.igem.org/Team:Cambridge#/Project/In_Vitro our wiki] for advice on protein purification and processing. | ||
| + | |||
| + | ====Characterisation of purified protein (in thin films)==== | ||
| + | |||
| + | ==Single Layer Spectra== | ||
| + | Thin films of polyHis-tagged reflectin A1 demonstrate rapid colour changes in response to vapour exposure (thin film swelling changes the interference patterns) | ||
| + | <gallery widths=300px heights=300px> | ||
| + | Image:Cam_Test1a.png | Contour plot of reflectance (height) against wavelength along the x-axis and time along the y-axis at the point where breathing was applied | ||
| + | Image:Cam_Test1b.png|Spectral data slices, they have been displaced in order to better illustrate the changes in profile with time with black indicating before breathing and red after | ||
| + | Image:Cam_Test1.png|Contour plot of reflectance (height) against wavelength along the x-axis and time along the y-axis for the entire capture duration showing the film reverts back to its original colouration | ||
| + | </gallery> | ||
| + | |||
| + | Looking at this we can see that by breathing we have been able to shift the colour of the film from blue to yellow. The presence of one dominant peak is an indication of the thickness of the film as we are only seeing one constructive interference determined by the dominant peak. | ||
| + | |||
| + | |||
| + | ====Characterisation ''in vivo''==== | ||
| + | |||
| + | This device was characterised ''in vivo'' as [[Part:BBa_K638301]], where reflectin is fused to GFP as an easy reporter for microscopy. We assessed folding on multiple backbones. | ||
| + | |||
| + | <gallery widths=400px heights=500px> | ||
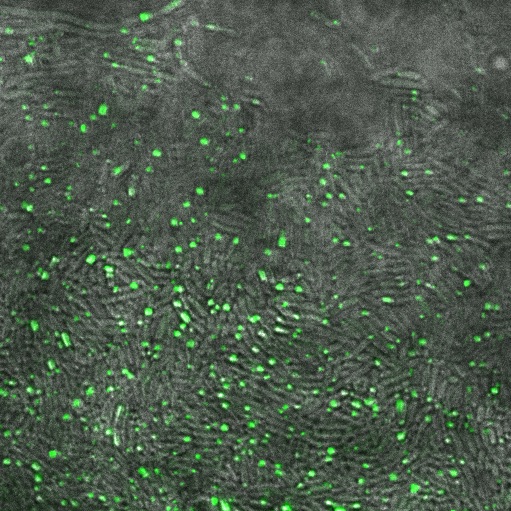
| + | Image:Cam_Reflectin-GFP-inclusionbodies.jpg | Reflectin-GFP on [[Part:pSB1A3 | pSB1A3]](a high copy plasmid), induced with 1mM Arabinose. Fluorescence is localised in dots at the end of the cells, indicating that fusion protein is localised to insoluble inclusion bodies. | ||
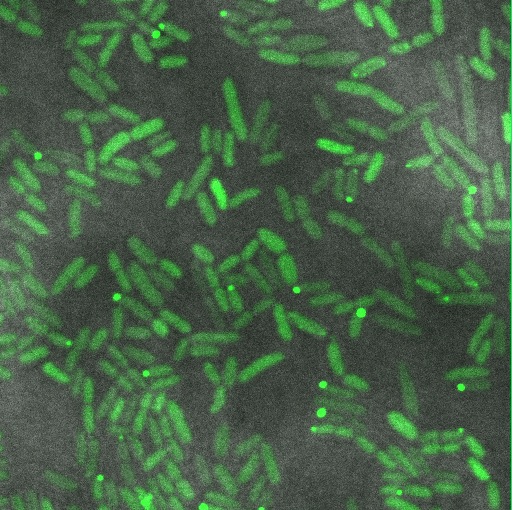
| + | Image:Cam-Low-copy-reflectin.jpg | Reflectin-GFP on [[Part:pSB3k3 | pSB3K3]] (low/med copy plasmid), induced with 1mM Arabinose. The fusion protein is distributed throughout the cytoplasm, indicating soluble protein is expressed. | ||
| + | </gallery> | ||
| + | |||
| + | From this confocal imaging, we recommend that reflectin be expressed on a '''low/med copy number backbone''' to allow protein folding to occur to give soluble protein. High copy number plasmids give inclusion bodies. Some inclusion bodies are also visible in the low copy construct, suggesting that some cells are expressing more fusion protein than others. See our [[Part:BBa_I0500:Experience | experience]] of the pBAD promoter for a discussion of variations of induction within a population of cells. | ||
| + | |||
| + | ====Safety==== | ||
| + | The protein coding sequence for reflectin originally came from cells of an edible squid. There have been no reported safety issues for reflectins, so we do not anticipate the need for extra precautions when using this BioBrick part. See our [http://2011.igem.org/Team:Cambridge/Safety safety page] for more information. | ||
Latest revision as of 00:45, 22 September 2011
Arabinose inducible Poly-His Reflectin A1 generator
Arabinose inducible Poly-His Reflectin A1 generator produces a poly-histidine (his-tagged) form of Reflectin A1( BBa_K638001).
Bacteria expressing this construct will produce Reflectin A1 with a poly-histidine tag at the N-terminus (upon induction with arabinose) in order to enable purification with a his-affinity column used in conjunction with nickel or cobalt resins.
The Reflectin A1 gene is under the control of the pBAD promoter BBa_I0500, which is tightly controlled by two factors:
- L-arabinose monosaccharide taken up by the cell from the medium, which acts as an inducer.
- AraC protein included in the I0500 biobrick, which acts an a repressor.
Therefore, the araC-pBAD system offers regulatable control of gene expression in the presence of the inducer and highly repressed in the absence of the inducer. Read more about [http://2011.igem.org/Team:Cambridge/Experiments/Low_Level_Expression the pBAD and arabinose system in E.coli].
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1205
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
Illegal BamHI site found at 1261 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961
Usage and Biology
Best used in an E. coli chassis such as strain [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783], with constitutive expression of an Arabinose transporter. See our experience of pBAD for some issues with tuning expression levels.
See [http://2011.igem.org/Team:Cambridge#/Project/In_Vitro our wiki] for advice on protein purification and processing.
Characterisation of purified protein (in thin films)
Single Layer Spectra
Thin films of polyHis-tagged reflectin A1 demonstrate rapid colour changes in response to vapour exposure (thin film swelling changes the interference patterns)
Looking at this we can see that by breathing we have been able to shift the colour of the film from blue to yellow. The presence of one dominant peak is an indication of the thickness of the film as we are only seeing one constructive interference determined by the dominant peak.
Characterisation in vivo
This device was characterised in vivo as Part:BBa_K638301, where reflectin is fused to GFP as an easy reporter for microscopy. We assessed folding on multiple backbones.
Reflectin-GFP on pSB1A3(a high copy plasmid), induced with 1mM Arabinose. Fluorescence is localised in dots at the end of the cells, indicating that fusion protein is localised to insoluble inclusion bodies.
Reflectin-GFP on pSB3K3 (low/med copy plasmid), induced with 1mM Arabinose. The fusion protein is distributed throughout the cytoplasm, indicating soluble protein is expressed.
From this confocal imaging, we recommend that reflectin be expressed on a low/med copy number backbone to allow protein folding to occur to give soluble protein. High copy number plasmids give inclusion bodies. Some inclusion bodies are also visible in the low copy construct, suggesting that some cells are expressing more fusion protein than others. See our experience of the pBAD promoter for a discussion of variations of induction within a population of cells.
Safety
The protein coding sequence for reflectin originally came from cells of an edible squid. There have been no reported safety issues for reflectins, so we do not anticipate the need for extra precautions when using this BioBrick part. See our [http://2011.igem.org/Team:Cambridge/Safety safety page] for more information.