Difference between revisions of "Part:BBa K638402"
(→Improving efficiency of export (Literature Data)) |
|||
| Line 2: | Line 2: | ||
<partinfo>BBa_K638402 short</partinfo> | <partinfo>BBa_K638402 short</partinfo> | ||
| − | This is an improved version of [https://parts.igem.org/Part:BBa_K233307 part BBa_K233307] designed to allow comparison with measurements of functionality in the literature, and to make it easier to synthesise. | + | This is an improved version of [https://parts.igem.org/Part:BBa_K233307 part BBa_K233307] designed to allow comparison with measurements of functionality in the literature, and to make it easier to synthesise. Length of the submitted part has been checked by colony PCR. |
| − | This TorA leader sequence variant has | + | This TorA leader sequence variant has been successfully used to export GFP to the periplasm of E.coli as described [http://www.ncbi.nlm.nih.gov/pubmed/11123687 here in the literature]. |
<!-- --> | <!-- --> | ||
Revision as of 15:30, 21 September 2011
TorA tag variant
This is an improved version of part BBa_K233307 designed to allow comparison with measurements of functionality in the literature, and to make it easier to synthesise. Length of the submitted part has been checked by colony PCR.
This TorA leader sequence variant has been successfully used to export GFP to the periplasm of E.coli as described [http://www.ncbi.nlm.nih.gov/pubmed/11123687 here in the literature].
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Improving efficiency of export (Literature Data)
[http://www.ncbi.nlm.nih.gov/pubmed/11123687 Thomas et al] described use of the part by fusing it to GFP.
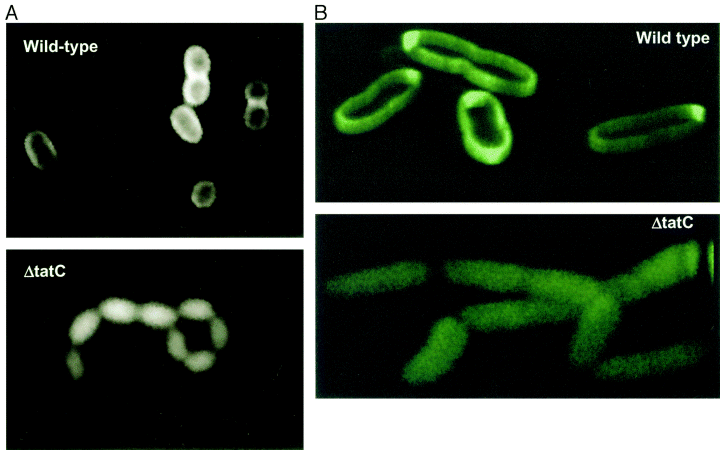
500px | Visualization of the periplasmic export of TorA–GFP by fluorescence microscopy. Wild-type MC4100AR cells (top) and the ΔtatC strain (bottom) containing the torA–GFP fusion plasmid pJDT1 were grown in the presence of arabinose, then resuspended in the absence of arabinose and incubated for 2.5 h. Image taken from [http://www.ncbi.nlm.nih.gov/pubmed/11123687 Thomas et al, 2001]
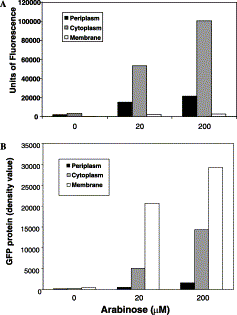
[http://www.ncbi.nlm.nih.gov/pubmed/12711311 Barrett et al] characterised improved export by altering growth conditions and upregulating TAT export machinery , using the same TorA-GFP fusion. They noted that efficiency of export is improved by removing induction from medium. As the below graph shows that the majority of GFP is membrane associated (and therefore does not fluoresce) - Barrett et al calculated that only ~4% of export tagged GFP may reach the periplasm in an active form when overexpressed.
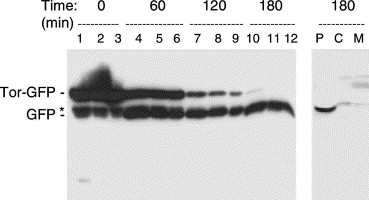
Marked increase in Tat-dependent export after removal of arabinose from the growth medium. Cells grown aerobically in lsLB and induced with 200 μM arabinose for 2 h at 37 °C. Cells were pelleted, washed with lsLB before being resuspended in lsLB. Samples of cells were removed and analysed immediately (T=0) or after 60, 120, or 180 min incubation with IPTG as indicated above the lanes. Samples containing equal numbers of cells were loaded on the gel (lanes 1, 4, 7, and 10), together with samples that were diluted 2-fold (lanes 2, 5, 8, and 11) and 4-fold (lanes 3, 6, 9, and 12) to aid comparison. The gel was immunoblotted with antibodies to GFP and the mobilities of TorA-GFP and mature GFP are indicated. Asterisk denotes apparent breakdown product of TorA-GFP that is present in the cytosol. Right hand lanes: a sample from the T=180 timepoint was fractionated to yield periplasmic (P), cytoplasmic (C), and membrane (M) samples and immunoblotted using antibodies to GFP. From Barrett et al, 2003.
Accumulated membrane-bound TorA-GFP is inactive. Upper graph: fractionated samples were assayed for fluorescence as detailed in Materials and methods. Values are given (in arbitrary units) for the fluorescence in the periplasmic (black), cytoplasmic (grey), and membrane (white) fractions. Lower graph shows values for the GFP levels in the same samples, calculated by quantitative immunoblotting; units are arbitrary. Barrett et al, 2003.
Producing BBa_k638402 from primer synthesis
We assembled the basic part by using 2 synthetic oligonucleotides (TorA-FWD and TorA-REV in the table and diagram below). These oligonucleotides have a 20bp overlap and can be used to generate the entire TorA tag by PCR.
| Name | Sequence | Tm |
|---|---|---|
| TorA FWD | ATGGCGAACAACGACTTATTTCAGGCTTCTCGGCGTCGCTTTCTGGCGCAGCTGGGCGGATTAACGGTGGCGGGT | 70.98°C |
| TorA REV | TGCGGCTTGTGCTGCCGTCGCTCTGCGAGGAGTCAACAGCGACGGGCCCAACATACCCGCCACCGTTAATCCGCC | 70.98°C |
Thermocycler profile: 10 cycles: Melt 95°C 10 sec / Anneal 65°C 30 sec / Extend 72°C 20 sec
Insertion of TorA tag into a construct
Cambridge 2011 created scar-free fusions of this export tag to our protein of interest by [http://www.cambridgeigem.org/RFC57.pdf Gibson Assembly]. Alternatively, Biobrick prefixes and suffixes could be added to this oligonucleotides (or added seperately) enabling the creation of in-frame fusions by assembly techniques such as BBF RFC 23 or 25.
The diagram below shows the 4 oligonucleotides needed to add a TorA tag by [http://www.cambridgeigem.org/RFC57.pdf Gibson Assembly]. TorA-FWD and TorA-REV generate the tag as described above. Gibson-primer-FWD and Gibson-primer-REV anneal to the template construct either side of where the TorA tag is to be placed. Gibson-primer-FWD has a tail which is the last 40bp of the TorA tag, and Gibson-primer-REV has a tail which is the reverse compliment of first 40bp of the TorA tag. These 40bp of overlap are what is required for [http://www.cambridgeigem.org/RFC57.pdf Gibson Assembly].