Difference between revisions of "Part:BBa K382002"
| (10 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K382002 short</partinfo> | <partinfo>BBa_K382002 short</partinfo> | ||
| − | Vector for transformation of plants. Cloning in E. Coli, transfer to agrobacteria, integrates in the plant genome. | + | Vector for transformation of plants. Cloning in ''E. Coli'', transfer to agrobacteria, integrates in the plant genome. Kanamycin resistance in bacteria, confers Basta resistance in plants. Contains the pENTCUP2 promoter upstream of MCS for constitutive expression of gene of interest.<br> |
| − | |||
| − | + | '''Confirmed functions''': | |
| + | * Kan resistance in bacteria: confirmed by transformation and colony growth of ''E. coli'' (DH5alpha) and ''Agrobacterium tumefaciens'' | ||
| + | * Basta resistance in plants: confirmed by selective germination of transformed ''Arabidopsis'' seeds on basta agar plates (See photos [http://2010.igem.org/Team:Harvard/results]).<br><br> | ||
| + | |||
| + | Confirmation pending: Function of the ENTCUP2 promoter will be tested by checking for expression for several BioBrick parts in adult ''Arabidopsis'' plants. These parts include... | ||
| + | * miraculin ([[Part:BBa_K382021|BBa_K382021]]) | ||
| + | * brazzein ([[Part:BBa_K382052|BBa_K382052]]) | ||
| + | * GFP knock-down amiRNA ([[Part:BBa_K382054|BBa_K382054]]) | ||
| + | |||
<br> | <br> | ||
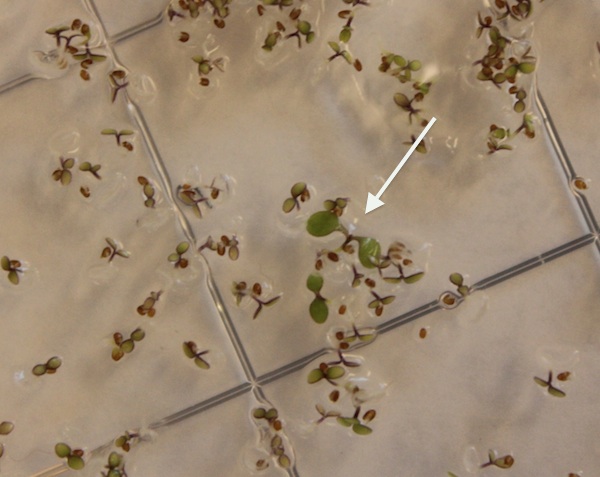
| + | Transformed seeds were plated on 5 micrograms/ml basta in Murashige and Skoog agar and we observed approximately 0.1% transgene expression and sprout survival. | ||
| + | |||
| + | [[Image:Bastaselection.jpg]] | ||
| + | <br> | ||
| + | Successfully transfected basta resistant ''Arabidopsis'' plants, such as the one shown below (brazzein [[Part:BBa_K382052|BBa_K382052]]), will be tested for BioBrick expression once they have grown large enough for tissue sampling: | ||
| + | <br> | ||
| + | |||
[[Image:BBa_K382002_plant.jpg|400px]] | [[Image:BBa_K382002_plant.jpg|400px]] | ||
| + | <br> | ||
| + | Download the annotated sequence file [http://openwetware.org/images/d/d2/V9.gb here]<br><br> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 17:18, 7 November 2010
pORE Expression Series Vector with BioBrick MCS, Basta resistance
Vector for transformation of plants. Cloning in E. Coli, transfer to agrobacteria, integrates in the plant genome. Kanamycin resistance in bacteria, confers Basta resistance in plants. Contains the pENTCUP2 promoter upstream of MCS for constitutive expression of gene of interest.
Confirmed functions:
- Kan resistance in bacteria: confirmed by transformation and colony growth of E. coli (DH5alpha) and Agrobacterium tumefaciens
- Basta resistance in plants: confirmed by selective germination of transformed Arabidopsis seeds on basta agar plates (See photos [http://2010.igem.org/Team:Harvard/results]).
Confirmation pending: Function of the ENTCUP2 promoter will be tested by checking for expression for several BioBrick parts in adult Arabidopsis plants. These parts include...
- miraculin (BBa_K382021)
- brazzein (BBa_K382052)
- GFP knock-down amiRNA (BBa_K382054)
Transformed seeds were plated on 5 micrograms/ml basta in Murashige and Skoog agar and we observed approximately 0.1% transgene expression and sprout survival.
Successfully transfected basta resistant Arabidopsis plants, such as the one shown below (brazzein BBa_K382052), will be tested for BioBrick expression once they have grown large enough for tissue sampling:
Download the annotated sequence file [http://openwetware.org/images/d/d2/V9.gb here]
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 5326
Illegal XbaI site found at 5299
Illegal SpeI site found at 5378
Illegal PstI site found at 5356 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 5326
Illegal NheI site found at 4766
Illegal SpeI site found at 5378
Illegal PstI site found at 5356
Illegal NotI site found at 5341 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 5326
Illegal BglII site found at 5212
Illegal BglII site found at 7771
Illegal BamHI site found at 5312
Illegal XhoI site found at 4790 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 5326
Illegal XbaI site found at 5299
Illegal SpeI site found at 5378
Illegal PstI site found at 5356 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 5326
Illegal XbaI site found at 5299
Illegal SpeI site found at 5378
Illegal PstI site found at 5356
Illegal NgoMIV site found at 625
Illegal NgoMIV site found at 749
Illegal NgoMIV site found at 2181
Illegal NgoMIV site found at 2772
Illegal NgoMIV site found at 7220
Illegal AgeI site found at 341 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 7241

