Difference between revisions of "Part:BBa K361011"
| (9 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K361011 short</partinfo> | <partinfo>BBa_K361011 short</partinfo> | ||
| − | This is the protein | + | This is the protein genertor of Rci site-specific recombinase BBa_K361000, for which a promotor and a ribosome binding site are linked on the left flank and a double bidirectional terminator on the right flank of the rci gene. |
| + | |||
| + | |||
| + | |||
| + | '''Performance'''<br> | ||
| + | <center> | ||
| + | {|{{Table}} | ||
| + | !Promotor | ||
| + | !lac promotor | ||
| + | |- | ||
| + | !RBS efficiency | ||
| + | !1 | ||
| + | |- | ||
| + | !Rci site specific recombinase | ||
| + | ![[Part:BBa_K361000|'''BBa_K361000''']] | ||
| + | |- | ||
| + | !terminator | ||
| + | !double bidirectional terminator | ||
| + | |} | ||
| + | </center> | ||
| + | |||
| + | Here is our system leading to the generation of Rci site-specific recombinase (yellow in colour) | ||
| + | [[Image:Rci_sys.png]]<br> | ||
| + | Fig. 1 - The structure of the Rci system | ||
| + | |||
| + | '''Validation'''<br> | ||
| + | In theory, with the use of repeat A at both ends of the [[Part:BBa_K361001|'''message''']] as shown in Fig. 2, there exists only two possible outcomes - a single inversion or double inversion. | ||
| + | [[Image:Our_message_structure.png|thumb|"Fig.2 - The structure of our message used in the validation process"]] | ||
| + | |||
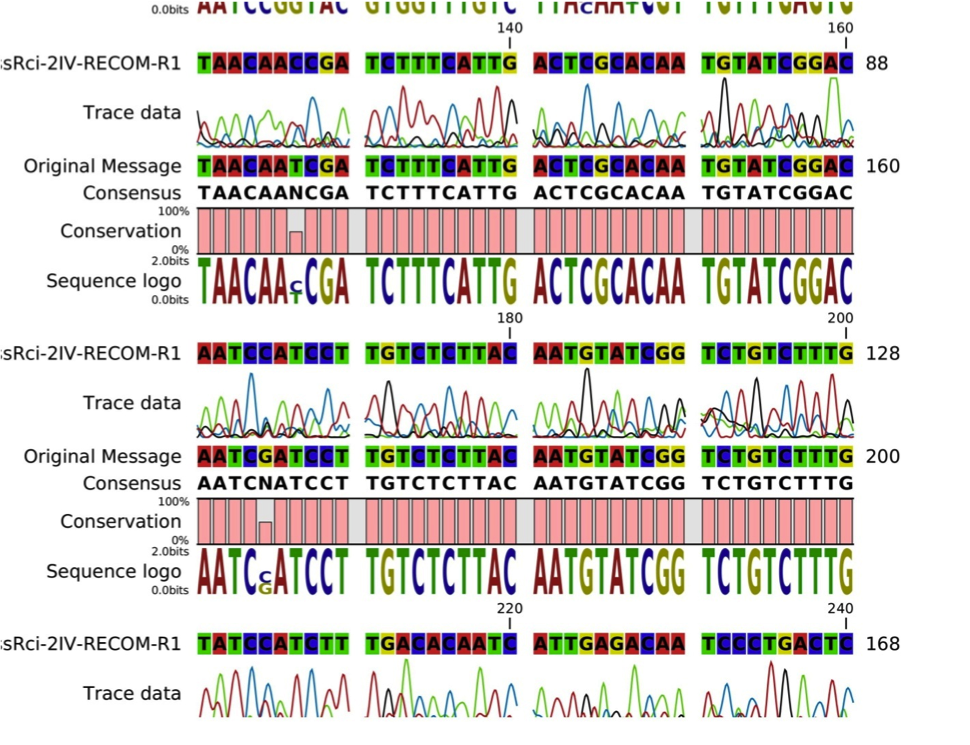
| + | With two sets of primers specifically designed for the amplification of both the single inversion product and double inversion product, the amplified DNA were then subjected to DNA sequencing. Both expected results are identified, however a few ambiguous bases were identified in the DNA sequencing process as shown in Fig. 3, this may due to the limitation of the sanger sequencing method or mutations along the growth of bacteria. | ||
| + | [[Image:Alignment_analysis.png|thumb|"Fig.3 - The alignment analysis of the sequenced results with the original sequence"]] | ||
| + | |||
| + | To conclude, since the majority of the sequence could be recovered, the Rci system was functioning correctly in our experiment. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 03:11, 7 November 2010
Rci recombinase system
This is the protein genertor of Rci site-specific recombinase BBa_K361000, for which a promotor and a ribosome binding site are linked on the left flank and a double bidirectional terminator on the right flank of the rci gene.
Performance
| Promotor | lac promotor |
|---|---|
| RBS efficiency | 1 |
| Rci site specific recombinase | BBa_K361000 |
| terminator | double bidirectional terminator |
Here is our system leading to the generation of Rci site-specific recombinase (yellow in colour)
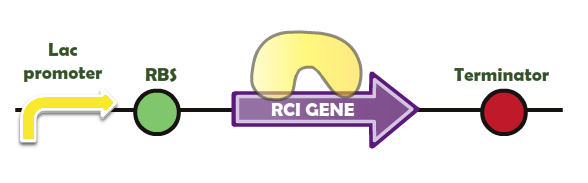
Fig. 1 - The structure of the Rci system
Validation
In theory, with the use of repeat A at both ends of the message as shown in Fig. 2, there exists only two possible outcomes - a single inversion or double inversion.
With two sets of primers specifically designed for the amplification of both the single inversion product and double inversion product, the amplified DNA were then subjected to DNA sequencing. Both expected results are identified, however a few ambiguous bases were identified in the DNA sequencing process as shown in Fig. 3, this may due to the limitation of the sanger sequencing method or mutations along the growth of bacteria.
To conclude, since the majority of the sequence could be recovered, the Rci system was functioning correctly in our experiment.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 588
Illegal NgoMIV site found at 1083 - 1000COMPATIBLE WITH RFC[1000]