Difference between revisions of "Part:BBa K404001"
| (10 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K404001 short</partinfo> | <partinfo>BBa_K404001 short</partinfo> | ||
| + | {| style="color:black; margin: 0px 0px 500px 20px;" cellpadding="6" cellspacing="1" border="2" align="right" | ||
| + | ! colspan="2" style="background:#66bbff;"|[https://parts.igem.org/Part:BBa_K404001 AAV2-Rep-VP123] | ||
| + | |- | ||
| + | |'''BioBrick Nr.''' | ||
| + | |[https://parts.igem.org/Part:BBa_K404001 BBa_K404001] | ||
| + | |- | ||
| + | |'''RFC standard''' | ||
| + | |[https://parts.igem.org/Help:Assembly_standard_25 RFC 25] | ||
| + | |- | ||
| + | |'''Requirement''' | ||
| + | |pSB1C3<br> | ||
| + | |- | ||
| + | |'''Source''' | ||
| + | |pAAV_RC from Stratagene | ||
| + | |- | ||
| + | |'''Submitted by''' | ||
| + | |[http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] | ||
| + | |} | ||
| + | <br><br> | ||
| + | <br><h2>Replication</h2><br> | ||
| + | |||
| + | [[Image:Freiburg10_Rep.png|thumb|left|480px]]<br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | The Adeno-associated virus (AAV) consists of two open reading frames (ORF), rep and cap ORF. The four non-structural rep genes are driven by two promoters located at map units 5 (p5 promoter) and 19 (p19 promoter). Rep proteins are involved in genome encapsidation, regulation of gene expression and replication of the viral genome. <br> | ||
| + | The two larger proteins Rep78/68 play an essential role in viral genome integration and regulation of AAV gene expression, whereas the smaller Rep proteins are involved in viral genome encapsidation. Rep proteins act both as repressors and activators of AAV transcription in respect to the absence and presence of helper viruses such as adenoviruses (Ad) or herpes simplex viruses (HSV) by interacting with several cellular proteins (Nash, Chen, Salganik, & Muzyczka, 2009).<br> | ||
| + | Furthermore, in the absence of Rep proteins, as it is the case in recombinant AAVs, integration of the viral genome into the human genome is rare and random. There are several hotspots for integration of wtAAV genomes such as the human chromosome 19q13.42, known as the AAVSI site, but as well some other accessible chromatin regions for preferred integration have been found (5p13.3 and 3p24.3). <br> | ||
| + | Integration into the human genome is mediated by the two regulatory proteins Rep68 and Rep78 driven by the AAV p5 promoter. The proteins bind to the Rep binding site (RBS) which is located within the inverted terminal repeats (ITRs). The minimal consensus Rep binding site (RBS) GAGT GAGC is found within the ITRs and in the p5 integration-efficient element (p5IEE) of the p5 promoter (Hüser et al., 2010). Rep78/68 proteins possess DNA-binding, helicase and site-specific endonuclease activity located within the first 200 amino acids (Davis, Wu, & Owens, 2000). Since the N-terminal region is unique to the larger Rep proteins, the two smaller Rep proteins possess other biological functions. <br> | ||
| + | Rep52/40 gene expression is driven by the p19 promoter which is located within rep ORF and the proteins are involved in encapsidating the viral genome into the preformed capsids.<br> | ||
| + | Gene expression of these proteins is suppressed in absence of adenovirus infection by binding of Rep78/68 to the p5 promoter. Gene expression of p19 and p40 is transacvtivated by the Rep proteins Rep78/68 during coinfection.<br><br> | ||
| + | '''Rep78'''<br> | ||
| + | Regulated by the p5 promoter, Rep78 is the largest non-structural protein found in the wtAAV. Besides regulation of gene expression and viral genome replication, Rep78 has been found to play a functional role in AAV site-specific integration into the human genome (Hüser et al., 2010). In absence of Ad helper viruses, overexpression of Rep78 leads to cell cycle arrest by interacting with cell-cycle regulating phosphatases causing DNA damage by its intrinsic endonuclease activity (Berthet, Raj, Saudan, & Beard, 2005) and induces apoptosis. Due to its ability to bind to the Rep binding site (RBS) in the p5 integration-efficient element (p5IEE) of the p5 promoter, Rep78 mediates gene expression and retain a constant level of Rep proteins by suppressing transcriptional activity of the p5 promoter in absence of Ad viruses (Yue et al., 2010). Interaction of Rep78 with cellular factors such as transcription factors (Lackner & Muzyczka, 2002) provides the basis for gene regulation by Rep78 in associated with endogenous molecules. | ||
| + | <br><br> | ||
| + | '''Rep68'''<br> | ||
| + | Rep68 is a regulatory protein driven by the p5 promoter with an apparent molecular weight of 68 kDa lacking 92 amino acids from the carboxy terminus due to splicing of mRNA coding for the two larger Rep proteins. | ||
| + | The non-structural protein Rep68 belongs to the superfamily 3 (SF3) helicase found in other small DNA and RNA viruses such as simian virus 40 (SV40) and bovine papillomavirus (Mansilla-Soto et al., 2009). Formation of oligomeric complexes of Rep proteins provides the basis for the functional versatility of the two larger regulatory proteins. The AAA+ motor domain is known to function as an initiator for oligomerization of the Rep proteins. The cooperative effect of both domains appears to be further regulated by ATP binding as well as different DNA substrates such as dsDNA and ssDNA. Assembly of different nucleoprotein structures suggest that viral replication and genome integration is regulated and controlled by distinct Rep complexes which means that in presence of dsDNA Rep68 assembles to smaller complexes than in presence of ssDNA resulting in octamers. | ||
| + | <br><br> | ||
| + | '''Rep52''' | ||
| + | Involved in genome encapsidation | ||
| + | Rep 52 is under the control of the p19 promoter and shares the same N-terminus with Rep78. It was shown that Rep52 possesses helicase and ATPase activity with 3´-5´polarity (Smith & Kotin, 1998). Despite the helicase activity, Rep52 and Rep78 share a putative zinc-finger domain, which suggest interactions with diverse cellular factors (Nash, Chen, Salganik, & Muzyczka, 2009) such as transcription factors (Lackner & Muzyczka, 2002) and TATA-binding proteins (Hermonat, Santin, Batchu, & Zhan, 1998). | ||
| + | <br><br> | ||
| + | '''Rep40'''<br> | ||
| + | The smallest Rep protein (Rep40) possesses helicase and ATPase activity as well, but does not have strict requirements for DNA duplexes containing a 3´single-stranded end. Rep40 helicase activity requires bivalent ions such as Mg2+ or Mn2+ and is most active using ATP as substrate. Lacking the zinc finger domain, present in Rep52, Rep40 requires dimerization for functional helicase activity (Collaco, Kalman-Maltese, Smith, Dignam, & Trempe, 2003). Rep40/52 proteins are required for translocation of the single-stranded, viral genomes into the preformed capsids proceeding with the 3´end of the DNA (King, Dubielzig, Grimm, & Kleinschmidt, 2001). | ||
| + | |||
| + | |||
<html> | <html> | ||
| − | < | + | <h2>Capsid</h2> |
| − | + | The AAV capsid consists of 60 capsid protein subunits. The three cap proteins VP1, VP2, and VP3 are encoded in an overlapping reading frame. Arranged in a stoichiometric ratio of 1:1:10, they form an icosahedral symmetry. The mRNA encoding for the cap proteins is transcribed from p40 and alternative spliced to minor and major products. Alternative splicing and translation initiation of VP2 at a nonconventional ACG initiation codon promote the expression of VP1, VP2 and VP3. The VP proteins share a common C terminus and stop codon, but begin with a different start codon. The N termini of VP1 and VP2 play important roles in infection and contain motifs that are highly homologous to a phospholipase A2 (PLA2) domain and nuclear localization signals (BR)(+). (Quelle) | |
| − | + | ||
| − | + | ||
| − | + | <center><img src="https://static.igem.org/mediawiki/parts/a/a7/Freiburg10_Cap_proteins_VP1_2%263.png" width="600" | |
| − | + | height="auto"/></center> | |
| − | + | <b> Figure 1: The VP proteins are encoded in an overlapping open reading frame. </b><br> | |
| − | + | <br> | |
| − | + | ||
| − | + | ||
| − | + | <html> | |
| − | + | <head> | |
| − | + | <meta content="text/html; charset=ISO-8859-1" | |
| − | + | http-equiv="content-type"> | |
| − | + | <title></title> | |
| − | + | </head> | |
| − | + | <body> | |
| − | + | <title>BBa_K404003</title> | |
| − | + | <br> | |
| − | + | <br><h2>Results</h2><br> | |
| − | + | <h3 style="margin-left: 0cm; text-indent: 0cm;"><a | |
| − | + | name="_Toc275885926"></a><a name="_Toc275817885"><span | |
| − | + | lang="EN-US">Modularization: Adapting | |
| − | + | pSB1C3 to loop | |
| − | + | insertions – pSB1C3_001</span></a></h3> | |
| − | + | <p class="MsoNormal" | |
| − | + | style="text-indent: 0cm; line-height: 150%;"><span | |
| − | + | lang="EN-US">To | |
| − | + | fulfill iGEM requirements, all plasmids need to be submitted in pSB1C3. | |
| − | + | Therefore, primers were ordered for amplifying <i>RepVP123</i> | |
| − | + | containing all | |
| − | + | modifications done so far by PCR and cloning them into pSB1C3. Still, | |
| − | + | pSB1C3 | |
| − | + | contains two restriction sites for SspI and PvuII restriction enzymes | |
| − | + | in its | |
| − | a | + | CAT marker. Since these are necessary for cloning ViralBricks in this |
| − | + | vector, | |
| − | + | the iGEM Team Freiburg_Bioware 2010 decided in agreement with iGEM | |
| − | + | Headquarters | |
| − | < | + | to implement a new standard for the pSB1C3 backbone which was named |
| − | < | + | pSB1C3_001. |
| − | + | Both restriction sites interfering with ViralBrick insertions were | |
| − | + | mutated to | |
| − | </ | + | make SspI and PvuII single-cutters (see method development).</span></p> |
| − | < | + | <table class="MsoTableGrid" |
| − | < | + | style="border: medium none ; margin-left: auto; border-collapse: collapse; text-align: left; margin-right: auto;" |
| − | </ | + | border="0" cellpadding="0" cellspacing="0"> |
| − | < | + | <tbody> |
| − | < | + | <tr> |
| − | < | + | <td |
| − | < | + | style="border: 1pt solid windowtext; padding: 0cm 5.4pt; vertical-align: top; width: 460.6pt;"> |
| − | < | + | <p class="MsoNormal" |
| − | < | + | style="text-align: center; text-indent: 0cm; page-break-after: avoid;" |
| − | </ | + | align="center"><img style="width: 584px; height: 278px;" |
| − | < | + | id="Picture 16" |
| − | + | src="https://static.igem.org/mediawiki/2010/d/d1/Freiburg10_pSB1C3_001_mutations.png" | |
| − | + | alt=""></p> | |
| − | </ | + | <p class="MsoCaption"><span lang="EN-US">Figure |
| − | < | + | 4 </span><span style="color: windowtext; font-weight: normal;" |
| − | < | + | lang="EN-US">Comparison |
| − | < | + | of pSB1C3 (upper row) and pSB1C3_001 (lower row). Deletions of SspI and |
| − | </ | + | PvuII are marked by red boxes.</span></p> |
| − | < | + | </td> |
| − | < | + | </tr> |
| − | < | + | </tbody> |
| − | <tr> | + | |
| − | <td | + | |
| − | < | + | |
| − | </tr> | + | |
</table> | </table> | ||
| − | </ | + | <p class="MsoNormal"><i><span lang="EN-US">RepVP123</span></i><span |
| − | + | lang="EN-US"> | |
| + | containing both <i>rep</i> and <i>cap</i> | ||
| + | synthetic gene fragments including | ||
| + | the re-mutation of KpnI and the downstream p5TATA-less promotor was | ||
| + | cloned into | ||
| + | the newly constructed pSB1C3_001. Testing this newly assembled plasmid | ||
| + | in cell | ||
| + | culture revealed unexpected data: Not only did the newly assembled | ||
| + | plasmid work | ||
| + | (see Figure 5), but in comparison to pAAV containing the same <i>RepVP123</i> | ||
| + | construct, pSB1C3_001 showed an about 3 times higher transduction | ||
| + | efficiency. | ||
| + | Although exact reasons are still unknown, these results are probably | ||
| + | related to | ||
| + | the length reduction of pSB1C3_001 compared to the original pAAV | ||
| + | plasmid | ||
| + | of | ||
| + | approximately 1000 base pairs.</span></p> | ||
| + | <span | ||
| + | style="font-size: 11pt; line-height: 200%; font-family: "Calibri","sans-serif";" | ||
| + | lang="EN-US"></span><span lang="EN-US"> </span> | ||
| + | <table | ||
| + | style="border: medium none ; width: 466px; border-collapse: collapse; text-align: left; margin-left: auto; margin-right: auto;" | ||
| + | class="MsoTableGrid" border="0" cellpadding="0" | ||
| + | cellspacing="0"> | ||
| + | <tbody> | ||
| + | <tr style="height: 209.85pt;"> | ||
| + | <td | ||
| + | style="border: 1pt solid windowtext; padding: 0cm 5.4pt; vertical-align: top; width: 4661px; height: 209.85pt;"> | ||
| + | <p class="MsoNormal" | ||
| + | style="text-align: center; text-indent: 0cm; page-break-after: avoid;" | ||
| + | align="center"><span | ||
| + | style="font-size: 10pt; line-height: 200%;"><img | ||
| + | style="width: 516px; height: 258px;" alt="" id="Chart 15" | ||
| + | src="https://static.igem.org/mediawiki/2010/2/2b/Freiburg10_pAAV_pSB1C3_001.png"></span></p> | ||
| + | <p class="MsoCaption"><span lang="EN-US">Figure | ||
| + | 5 </span><span style="color: windowtext; font-weight: normal;" | ||
| + | lang="EN-US">AAV-293 | ||
| + | cells were transfected with three plasmids pHelper, | ||
| + | pSB1C3_001_[AAV2]-Rep-VP123_p5-TATAless or pAAV_RC_IRCK and | ||
| + | pSB1C3_[AAV2]-left-ITR_pCMV_beta-globin_mVenus_hGH_[AAV2]-right-ITR | ||
| + | providing essential genes and proteins for producing viral particles. | ||
| + | 48 hours post transfection, viral particles were harvested by | ||
| + | freeze-thaw lysis and centrifugation followed by HT1080 transduction. | ||
| + | mVenus expression of viral genomes was determined by flow cytometry | ||
| + | analysis | ||
| + | 24 hours post infection. </span><span | ||
| + | style="color: windowtext; font-weight: normal;" lang="EN-US">Fluorescence | ||
| + | is measured in surviving cells.</span><span | ||
| + | style="color: windowtext; font-weight: normal;" lang="EN-US"> Results | ||
| + | showed functionality of <i>RepVP123</i> | ||
| + | within the pSB1C3_001 vector and additionally increased transduction | ||
| + | efficiency.</span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <h4 style="margin-left: 0cm; text-indent: 0cm;"><a | ||
| + | name="_Toc275885927"></a><a name="_Toc275817886"><span | ||
| + | lang="EN-US"></span></a></h4> | ||
| + | </body> | ||
| + | |||
| + | |||
| + | <h3>References</h3> | ||
| + | <b>Cassinotti, P., Weitzand, M., & Tratschin, J. D.</b>, 1988. Organization of the adeno-associated virus (AAV) capsid gene: mapping of a minor spliced mRNA coding for virus capsid protein. Virology, 167(1), 176-84 <br /> | ||
| + | |||
| + | |||
| + | </html> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 15:27, 30 October 2010
[AAV2]-Rep-VP123
| AAV2-Rep-VP123 | |
|---|---|
| BioBrick Nr. | BBa_K404001 |
| RFC standard | RFC 25 |
| Requirement | pSB1C3 |
| Source | pAAV_RC from Stratagene |
| Submitted by | [http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] |
Replication
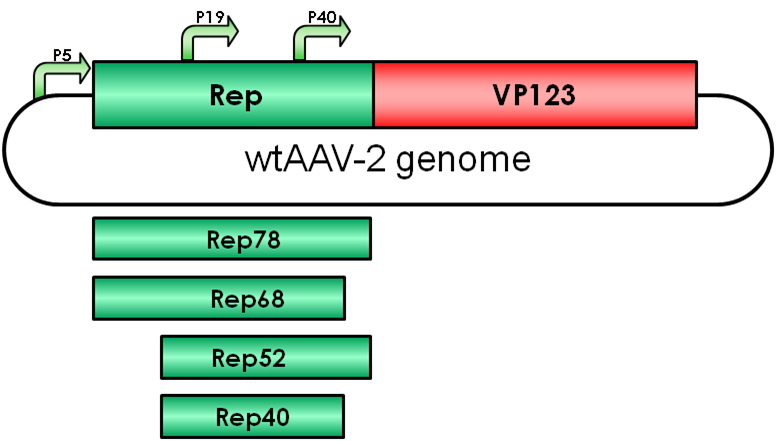
The Adeno-associated virus (AAV) consists of two open reading frames (ORF), rep and cap ORF. The four non-structural rep genes are driven by two promoters located at map units 5 (p5 promoter) and 19 (p19 promoter). Rep proteins are involved in genome encapsidation, regulation of gene expression and replication of the viral genome.
The two larger proteins Rep78/68 play an essential role in viral genome integration and regulation of AAV gene expression, whereas the smaller Rep proteins are involved in viral genome encapsidation. Rep proteins act both as repressors and activators of AAV transcription in respect to the absence and presence of helper viruses such as adenoviruses (Ad) or herpes simplex viruses (HSV) by interacting with several cellular proteins (Nash, Chen, Salganik, & Muzyczka, 2009).
Furthermore, in the absence of Rep proteins, as it is the case in recombinant AAVs, integration of the viral genome into the human genome is rare and random. There are several hotspots for integration of wtAAV genomes such as the human chromosome 19q13.42, known as the AAVSI site, but as well some other accessible chromatin regions for preferred integration have been found (5p13.3 and 3p24.3).
Integration into the human genome is mediated by the two regulatory proteins Rep68 and Rep78 driven by the AAV p5 promoter. The proteins bind to the Rep binding site (RBS) which is located within the inverted terminal repeats (ITRs). The minimal consensus Rep binding site (RBS) GAGT GAGC is found within the ITRs and in the p5 integration-efficient element (p5IEE) of the p5 promoter (Hüser et al., 2010). Rep78/68 proteins possess DNA-binding, helicase and site-specific endonuclease activity located within the first 200 amino acids (Davis, Wu, & Owens, 2000). Since the N-terminal region is unique to the larger Rep proteins, the two smaller Rep proteins possess other biological functions.
Rep52/40 gene expression is driven by the p19 promoter which is located within rep ORF and the proteins are involved in encapsidating the viral genome into the preformed capsids.
Gene expression of these proteins is suppressed in absence of adenovirus infection by binding of Rep78/68 to the p5 promoter. Gene expression of p19 and p40 is transacvtivated by the Rep proteins Rep78/68 during coinfection.
Rep78
Regulated by the p5 promoter, Rep78 is the largest non-structural protein found in the wtAAV. Besides regulation of gene expression and viral genome replication, Rep78 has been found to play a functional role in AAV site-specific integration into the human genome (Hüser et al., 2010). In absence of Ad helper viruses, overexpression of Rep78 leads to cell cycle arrest by interacting with cell-cycle regulating phosphatases causing DNA damage by its intrinsic endonuclease activity (Berthet, Raj, Saudan, & Beard, 2005) and induces apoptosis. Due to its ability to bind to the Rep binding site (RBS) in the p5 integration-efficient element (p5IEE) of the p5 promoter, Rep78 mediates gene expression and retain a constant level of Rep proteins by suppressing transcriptional activity of the p5 promoter in absence of Ad viruses (Yue et al., 2010). Interaction of Rep78 with cellular factors such as transcription factors (Lackner & Muzyczka, 2002) provides the basis for gene regulation by Rep78 in associated with endogenous molecules.
Rep68
Rep68 is a regulatory protein driven by the p5 promoter with an apparent molecular weight of 68 kDa lacking 92 amino acids from the carboxy terminus due to splicing of mRNA coding for the two larger Rep proteins.
The non-structural protein Rep68 belongs to the superfamily 3 (SF3) helicase found in other small DNA and RNA viruses such as simian virus 40 (SV40) and bovine papillomavirus (Mansilla-Soto et al., 2009). Formation of oligomeric complexes of Rep proteins provides the basis for the functional versatility of the two larger regulatory proteins. The AAA+ motor domain is known to function as an initiator for oligomerization of the Rep proteins. The cooperative effect of both domains appears to be further regulated by ATP binding as well as different DNA substrates such as dsDNA and ssDNA. Assembly of different nucleoprotein structures suggest that viral replication and genome integration is regulated and controlled by distinct Rep complexes which means that in presence of dsDNA Rep68 assembles to smaller complexes than in presence of ssDNA resulting in octamers.
Rep52
Involved in genome encapsidation
Rep 52 is under the control of the p19 promoter and shares the same N-terminus with Rep78. It was shown that Rep52 possesses helicase and ATPase activity with 3´-5´polarity (Smith & Kotin, 1998). Despite the helicase activity, Rep52 and Rep78 share a putative zinc-finger domain, which suggest interactions with diverse cellular factors (Nash, Chen, Salganik, & Muzyczka, 2009) such as transcription factors (Lackner & Muzyczka, 2002) and TATA-binding proteins (Hermonat, Santin, Batchu, & Zhan, 1998).
Rep40
The smallest Rep protein (Rep40) possesses helicase and ATPase activity as well, but does not have strict requirements for DNA duplexes containing a 3´single-stranded end. Rep40 helicase activity requires bivalent ions such as Mg2+ or Mn2+ and is most active using ATP as substrate. Lacking the zinc finger domain, present in Rep52, Rep40 requires dimerization for functional helicase activity (Collaco, Kalman-Maltese, Smith, Dignam, & Trempe, 2003). Rep40/52 proteins are required for translocation of the single-stranded, viral genomes into the preformed capsids proceeding with the 3´end of the DNA (King, Dubielzig, Grimm, & Kleinschmidt, 2001).
Capsid
The AAV capsid consists of 60 capsid protein subunits. The three cap proteins VP1, VP2, and VP3 are encoded in an overlapping reading frame. Arranged in a stoichiometric ratio of 1:1:10, they form an icosahedral symmetry. The mRNA encoding for the cap proteins is transcribed from p40 and alternative spliced to minor and major products. Alternative splicing and translation initiation of VP2 at a nonconventional ACG initiation codon promote the expression of VP1, VP2 and VP3. The VP proteins share a common C terminus and stop codon, but begin with a different start codon. The N termini of VP1 and VP2 play important roles in infection and contain motifs that are highly homologous to a phospholipase A2 (PLA2) domain and nuclear localization signals (BR)(+). (Quelle)
Results
Modularization: Adapting pSB1C3 to loop insertions – pSB1C3_001
To fulfill iGEM requirements, all plasmids need to be submitted in pSB1C3. Therefore, primers were ordered for amplifying RepVP123 containing all modifications done so far by PCR and cloning them into pSB1C3. Still, pSB1C3 contains two restriction sites for SspI and PvuII restriction enzymes in its CAT marker. Since these are necessary for cloning ViralBricks in this vector, the iGEM Team Freiburg_Bioware 2010 decided in agreement with iGEM Headquarters to implement a new standard for the pSB1C3 backbone which was named pSB1C3_001. Both restriction sites interfering with ViralBrick insertions were mutated to make SspI and PvuII single-cutters (see method development).
|
Figure 4 Comparison of pSB1C3 (upper row) and pSB1C3_001 (lower row). Deletions of SspI and PvuII are marked by red boxes. |
RepVP123 containing both rep and cap synthetic gene fragments including the re-mutation of KpnI and the downstream p5TATA-less promotor was cloned into the newly constructed pSB1C3_001. Testing this newly assembled plasmid in cell culture revealed unexpected data: Not only did the newly assembled plasmid work (see Figure 5), but in comparison to pAAV containing the same RepVP123 construct, pSB1C3_001 showed an about 3 times higher transduction efficiency. Although exact reasons are still unknown, these results are probably related to the length reduction of pSB1C3_001 compared to the original pAAV plasmid of approximately 1000 base pairs.
|
Figure 5 AAV-293 cells were transfected with three plasmids pHelper, pSB1C3_001_[AAV2]-Rep-VP123_p5-TATAless or pAAV_RC_IRCK and pSB1C3_[AAV2]-left-ITR_pCMV_beta-globin_mVenus_hGH_[AAV2]-right-ITR providing essential genes and proteins for producing viral particles. 48 hours post transfection, viral particles were harvested by freeze-thaw lysis and centrifugation followed by HT1080 transduction. mVenus expression of viral genomes was determined by flow cytometry analysis 24 hours post infection. Fluorescence is measured in surviving cells. Results showed functionality of RepVP123 within the pSB1C3_001 vector and additionally increased transduction efficiency. |
References
Cassinotti, P., Weitzand, M., & Tratschin, J. D., 1988. Organization of the adeno-associated virus (AAV) capsid gene: mapping of a minor spliced mRNA coding for virus capsid protein. Virology, 167(1), 176-84Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 3611
Illegal XhoI site found at 1913
Illegal XhoI site found at 2099 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 4137
Illegal SapI site found at 3048