Part:BBa_K3740042
Chimeric Bacteriocin SE
Description
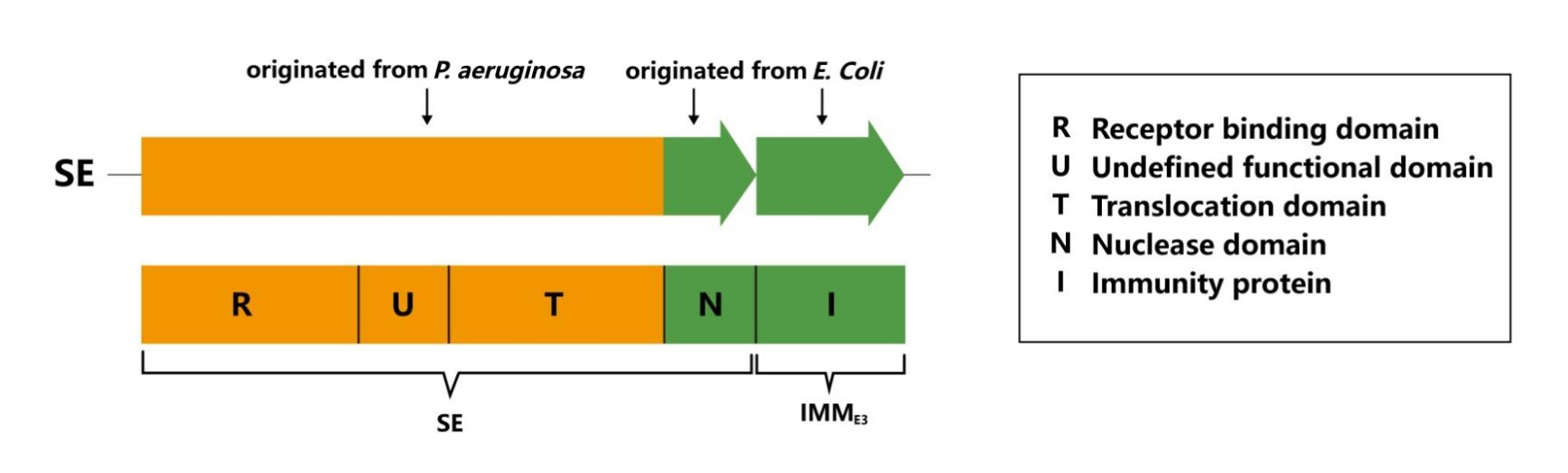
We replaced the nuclease domain of pyocin S2 with that of colicin E3 to produce a chimeric bacteriocin termed SE.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 2407
- 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 2407
- 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 2407
Illegal BglII site found at 2112
Illegal BamHI site found at 1684 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 2407
- 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 2407
Illegal NgoMIV site found at 1483
Illegal AgeI site found at 1573 - 1000COMPATIBLE WITH RFC[1000]
2021 SZPT-China
Biology
The receptor domain, translocase domain of pyocin S2 and the nuclease domain of colicin E3 are fused together to get a new chimeric bacteriocin termed SE. The antipseudomonal SE protein can target and inhibit P. aeruginosa, even including those harbouring a S2 immunity protein IMMS2.
Usage
The chimeric bacteriocin SE (BBa_K3740042) was constructed with an inducible expression vector to form the SE-PET28A expression vector, and transferred into E. coli BL21 to verify its function.
Characterization
1.SDS-PAGE
Method: IPTG was added to a final concentration of 0.1 mmol/mL to induce SE protein expression in E. coli BL21 and subsequently incubated at 25℃ for 12 hours. Supernatant containing SE or S2 proteins was obtained by sonication and centrifugation respectively. The crude protein extract was loaded into SDS-PAGE gel and stained by Coomassie Blue to verify the presence of target proteins.
After induction by IPTG, S2 and SE protein expression in S2-PET28A-BL21 (lane 2) and SE-PET28A-BL21 (lane 3) has been identified. By contrast, target proteins were not expressed in the strain with S2-PET28A-BL21 (lane 1) and SE -PET28A-BL21 (lane 4) in the absence of IPTG, indicating that expression of S2 and SE proteins can be successfully induced in E. coli transformed with S2-PET28A and SE-PET28A, respectively.
2. SE protein targeting verification
Method: Supernatant containing SE or S2 protein was respectively mixed with the culture of Pseudomonas aeruginosa PAO1, P. aeruginosa PAO1Δ1150-1151(PAO1 Δ1150-1151, PAO1 knocked out of S2+immS2) and E. coli MG1655. PBS was used as a negative control. Inhibitory effect of supernatant containing SE or S2 protein on the P. aeruginosa growth was determined by measuring OD600 using SYNERGY H1 micro-plate reader.
We found that the OD600 values of both P. aeruginosa strains cultured with SE protein, were much lower than that of the control group PBS. This proves that SE protein has antipseudomonal properties. In contrast, pyocin S2 could only act on the immune-deficiency strain PAO1∆1150-1151. This proves that SE protein has a broader antipseudomonal spectrum than S2. Meanwhile, the OD600 value of E. coli MG1655 cultured with SE protein was a little higher than that of the control group PBS. This indicates that SE protein does not work on E. coli. This result indicated that the fusion protein SE can specifically kill P. aeruginosa.
References
[1] Gupta S, Bram E E, Weiss R. Genetically programmable pathogen sense and destroy. [J]. Acs Synthetic Biology, 2013, 2(12):715-723
| None |