File list
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | Description | Versions |
|---|---|---|---|---|---|
| 19:39, 6 April 2012 | KAH010812.jpg (file) |  |
90 KB | 1 | |
| 21:36, 5 April 2012 | Haynes mCherry PDB2H5Q.png (file) | 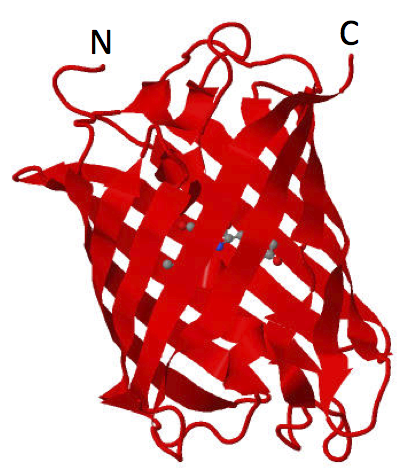 |
209 KB | mCherry Jmol rendering, PDB 2H5Q | 1 |
| 21:32, 5 April 2012 | Haynes discosomasp.png (file) |  |
317 KB | Discosoma sp. | 1 |
| 20:42, 5 April 2012 | Haynes Pc1 PDB1PDQ.png (file) |  |
89 KB | Jmol rendering of the 3D structure of the Drosophila Polycomb chromodomain | 1 |
| 20:42, 5 April 2012 | Haynes CBX8 PDB3I91.png (file) |  |
106 KB | Jmol rendering of the 3D structure of the human Polycomb chromodomain | 1 |
| 03:46, 8 February 2012 | Haynes Promoters mCh.tiff (file) | 193 KB | Microscopy of transiently transfected U2OS cells expressing BioBrick assembled human promoter-driven mCherry. The data shown here have not been published elsewhere. | 2 | |
| 03:43, 8 February 2012 | Haynes Promoters mCh.png (file) | 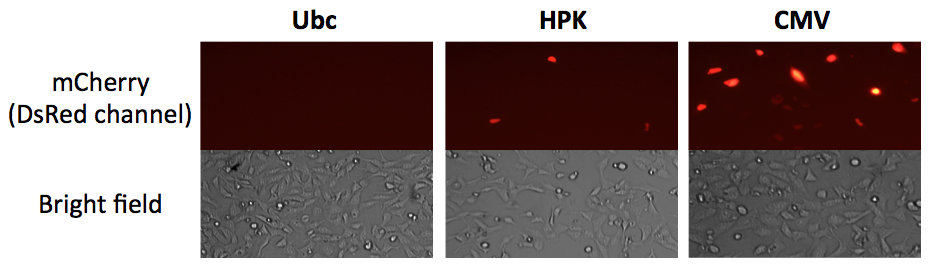 |
197 KB | Microscopy of transiently transfected U2OS cells expressing BioBrick assembled human promoter-driven mCherry. The data shown here have not been published elsewhere. | 1 |
| 17:49, 22 November 2011 | KAH VP64.jpg (file) |  |
21 KB | 1 | |
| 03:16, 22 November 2011 | KAH VP64.png (file) |  |
24 KB | 1 | |
| 05:09, 3 July 2011 | PCD Fischle 2003.png (file) |  |
50 KB | 1 | |
| 05:01, 3 July 2011 | PCD Fischle 2003.tif (file) | 56 KB | Crystal structure of Drosophila melanogaster Pc Polycomb chromodomain. Figure 6 from Fischle et al., 2003, Genes Dev. | 1 | |
| 18:24, 29 October 2006 | Hin dimer closed 1GDT.gif (file) |  |
28 KB | A Hin dimer bound to a HixC site. Open nicks in the DNA backbone show where the plus and minus strands have been cut by Hin. Protein Data Bank ID 1GDT. Yang, W., Steitz, T.A. (1995) ''Crystal structure of the site-specific recombinase gamma delta resolv | 1 |
| 18:13, 29 October 2006 | Hin dimer cut 2GM4.gif (file) |  |
27 KB | A Hin protein dimer bound to cleaved DNA. Protein Data Bank ID 2GM4. Kamtekar, S., Ho, R.S., Cocco, M.J., Li, W., Wenwieser, S.V.C.T., Boocock, M.R., Grindley, N.D.F., Steitz, T.A. (2006) ''An activated, tetrameric gamma-delta resolvase: Hin chim | 1 |
| 16:46, 29 October 2006 | NheI map.gif (file) |  |
32 KB | NheI restriction site map of two constructs ("pBad" and "Tet") which carry an invertable (HixC-flanked) segment of DNA. In the "pBad" construct, when the pBad promoter (green bent arrow) is flipped, the restriction fragment changes from 200 bp to 300 bp. | 2 |
| 03:56, 29 October 2006 | Tet flipping.jpg (file) |  |
8 KB | 1 | |
| 03:56, 29 October 2006 | PBad flipping.jpg (file) |  |
11 KB | 1 | |
| 03:52, 29 October 2006 | Jmol Hin tetrad DNA.gif (file) |  |
42 KB | Protein Data Bank ID 1ZR4, Jmol image. A Hin protein dimer binds and cleaves DNA at each HixC sequence flanking the fragment of DNA to be inverted. The two dimers (dimer 1 = leftward green and blue protein structures; dimer 2 = rightward yellow and purple | 1 |
