Recombination/Bacteriophage lambda-derived att
< Back to Recombination systems, DNA recombination sites, or Recombinases
| The 2004 Boston University iGEM team designed and constructed the att recombination sites BBa_I11022 and BBa_I11023. |
The following is excerpted from Radman-Livaja et al. RadmanLivaja and Landy et al. Landy. It has been edited for clarity.
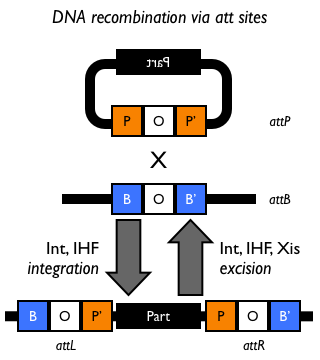
Bacteriophage λ has long served as a model system for studies of regulated site-specific recombination. In conditions favorable for bacterial growth, the phage genome is inserted into the Escherichia coli genome by an ‘integrative’ recombination reaction, which takes place between DNA attachment sites called attP and attB in the phage and bacterial genomes, respectively. As a result, the integrated λ DNA is bounded by hybrid attachment sites, termed attL and attR. In response to the physiological state of the bacterial host or to DNA damage, λ phage DNA excises itself from the host chromosome. This excision reaction recombines attL with attR to precisely restore the attP and attB sites on the circular λ and E. coli DNAs Campbell. The phage-encoded λ integrase protein (Int), a tyrosine recombinase, splices together bacterial and phage attachment sites. Int is required for both integration and excision of the λ prophage Zissler67.
λ recombination has a strong directional bias in response to environmental conditions. Accessory factors, whose expression levels change in response to host physiology, control the action of Int and determine whether the phage genome will remain integrated or be excised. Int has two DNA-binding domains: a C-terminal domain, consisting of a catalytic domain and a core-binding (CB) domain, that interacts with the core recombining sites and an N-terminal domain (N-domain) that recognizes the regulatory arm DNA sites Wojciak02. The heterobivalent Int molecules bridge distant core and arm sites with the help of accessory proteins, such as integration host factor (IHF), which bend the DNA at intervening sites, and appose arm and core sequences for interaction with the Int recombinase. Five arm DNA sites in the regions flanking the core of attP are differentially occupied during integration and excision reactions. The integration products attL and attR cannot revert back to attP and attB without assistance from the phage-encoded factor Xis, which bends DNA on its own or in combination with the host-encoded factor Fis Abremski82,Abremski81,Hoess80,Thompson87a,Thompson87b,Ball91. Xis also inhibits integration, and prevents the attP and attB products of excision from reverting to attL and attR Abremski81,18]. Excision is inhibited by high concentrations of IHF <cite>Bushman85,Thompson86. Because the cellular levels of IHF and Fis proteins respond to growth conditions, these host-encoded factors have been proposed as the master signals for integration and excision Thompson87a,Bushman85,Thompson86, Nilsson92,Ball92, Ball91.
The phage (attP) and bacterial (attB) att sites are designated POP’ and BOB’, respectively, and the prophage att sites are designated BOP’ (attL) and POB’ (attR).
Recombination sites
| Name | Description | Sequence | Recombinase | Length |
|---|---|---|---|---|
| BBa_I11022 | Lambda attB, reverse complement | accactttgtacaagaaagctgggt | 25 | |
| BBa_I11023 | Lambda attP | . . . tcactatcagtcaaaataaaatcattattt | 232 | |
| BBa_K112141 | attR2 recombination site | . . . gttcagctttcttgtacaaagtggttgatc | 136 | |
| BBa_K112142 | attR2 recombination site-reverse orientation | . . . aacacaacatatccagtcactatggtcgac | 136 |
Recombinases
| Name | Protein | Description | Direction | KEGG | UniProt | E.C. | Recombination site | Length |
|---|---|---|---|---|---|---|---|---|
| BBa_I11021 | Xis lambda | excisionase from E. coli phage lambda (removes prophage from host genome) | Forward | none | P03699 | none | 280 | |
| BBa_I11020 | Int lambda | integrase from E. coli phage lambda | Forward | none | P03700 | none | 1132 | |
| BBa_K112001 | Xis | Xis from bacteriophage lambda, assembly standard 21 | 216 | |||||
| BBa_K112204 | {a~xis!} The bacteriophage lambda xis gene ready to have rbs attached and stop codon; assembly stand | 223 | ||||||
| BBa_K112200 | {xis!} from bacteriophage lambda; assembly standard 21 | 219 |
Composite parts
| Name | Type | Description | Length | Status |
|---|---|---|---|---|
| BBa_K112137 | Composite | Xis Int & TrfA combined expression cassette | 2712 | It's complicated |
| BBa_K112132 | Composite | Xis Int controlled by temperature sensitive | 2169 | It's complicated |
| BBa_K112133 | Composite | Xis Int temperature sensitive expression cassette | 2215 | It's complicated |
| BBa_K112327 | Protein_Domain | {rbs_pelB>} {< xis>} | 304 | Not in stock |
References
<biblio>
- Campbell Campbell, AM. Episomes. In: Caspari EW, Thoday JM. , editors. Advances in Genetics. 1. New York: Academic Press; 1962. pp. 101–145.
- Zissler67 pmid=5637199
- Guameros70 pmid=4907272
- Kaiser70 pmid=4907271
- Echols70 pmid=4907273
- Shimada72 pmid=4552408
- Shimada75 pmid=1095763
- Fiandt72 pmid=4567155
- Hirsch72 pmid=4567154
- Hoess80 pmid=6446713
- Abremski81 pmid=6279866
- Abremski82 pmid=6213611
- Bushman85 pmid=2932798
- Thompson86 pmid=2946666
- Thompson87a pmid=2957063
- Thompson87b pmid=2958633
- Kitts88a pmid=2970060
- Kitts88b pmid=2975338
- NunesDuby87 pmid=3040260
- Holliday64 Holliday, R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304.
- Ball91 pmid=1829453
- Ball92 pmid=1459953
- Nilsson92 pmid=1732224
- Hallett97 pmid=9348666
- Azaro02 Azaro, MA; Landy, A. λ Int and the λ Int family. In: Craig NL, Craigie R, Gellert M, Lambowitz A. , editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 118–148.
- Wojciak02 pmid=11904406
- Landy pmid=331474
- Landy89 pmid=2528323
- RadmanLivaja pmid=16368232
</biblio>