Part:BBa_K808003
tctB_162: small subunit B1 of the tripartite tricarboxylate transporter family
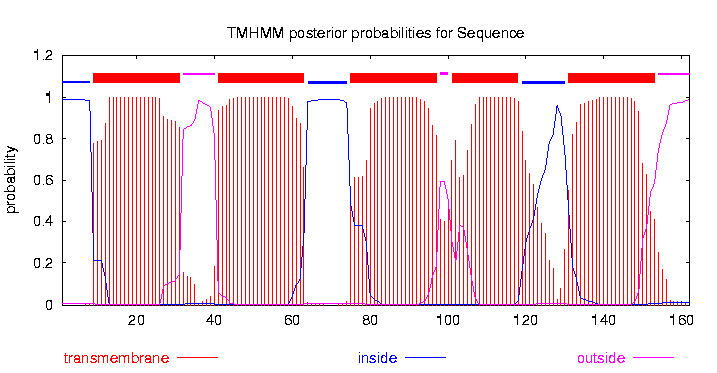
The small subunit B1 of the tripartite tricarboxylate transporter family (tctB_162, 17 kDa) was isolated from Comamonas testosteroni KF-1. The tripartite tricarboxylate transporter system consists of three different proteins: a periplasmatic solute binding receptor, a membrane protein with 12 putative transmembrane alpha-helical spanners (in this case tctB_162), and a small poorly conserved membrane proteine with four putative transmembrane alpha-helical spanners[1].The strain was purchased from Leibniz Institute DMSZ-German Collection of Microorganism and Cell Cultures (DMSZ no. 14576). The original sequence contains a Pst1 recognition site. To eliminate this recognition site a directed-site mutagenic PCR was performed. (For more datails:[http://2012.igem.org/Team:TU_Darmstadt/Protocols/mutagenic_PCR mutagenic PCR)] To characterized the structure of the tctB_162 bioinformatic tools like P rotein H omology/anolog Y R ecognition E ngine V 2.0 (PHYRE2), I-TASSER servers, protein B asic L ocal A ligment S earch T ool (BLAST) and TMHMM was used. The TMHMM predicted a transmembrane protein with 5 alpha-helical spanners (Fig. 1). The N-teminus is with a probability of over 99 % in cytoplasmatic. The NCBI Protein BLAST results shows that the tctB_162 subunit B1 belongs to the tctB superfamily.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 291
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 55
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
References
[1] Sasoh, M., E. Masai, et al. (2006). "Characterization of the terephthalate degradation genes of Comamonas sp. strain E6." Appl Environ Microbiol 72(3): 1825-1832.
- Fukuhara, Y., K. Inakazu, et al. (2010). "Characterization of the isophthalate degradation genes of Comamonas sp. strain E6." Appl Environ Microbiol 76(2): 519-527.
- Kamimura, N., T. Aoyama, et al. (2010). "Characterization of the protocatechuate 4,5-cleavage pathway operon in Comamonas sp. strain E6 and discovery of a novel pathway gene." Appl Environ Microbiol 76(24): 8093-8101.
- Winnen, B., R. N. Hvorup, et al. (2003). "The tripartite tricarboxylate transporter (TTT) family." Res Microbiol 154(7): 457-465.
- Protein structure prediction on the web: a case study using the PhyreKelley LA and Sternberg MJE.Nature Protocols 4, 363 - 371 (2009 server
- Yang Zhang. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, vol 9, 40 (2008).
- Ambrish Roy, Alper Kucukural, Yang Zhang. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, vol 5, 725-738 (2010).
- Ambrish Roy, Jianyi Yang, Yang Zhang. COFACTOR: An accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Research, doi:10.1093/nar/gks372 (2012)
- Prediction of twin-arginine signal peptides. Jannick Dyrløv Bendtsen, Henrik Nielsen, David Widdick, Tracy Palmer and Søren Brunak. BMC bioinformatics 2005 6: 167.
- SignalP 4.0: discriminating signal peptides from transmembrane regions Thomas Nordahl Petersen, Søren Brunak, Gunnar von Heijne & Henrik Nielsen Nature Methods, 8:785-786, 2011
- Erik L.L. Sonnhammer, Gunnar von Heijne, and Anders Krogh:A hidden Markov model for predicting transmembrane helices in protein sequences. In Proc. of Sixth Int. Conf. on Intelligent Systems for Molecular Biology, p 175-182,Ed J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen Menlo Park, CA: AAAI Press, 1998
| None |