Help:Terminators/Measurement/Jason Kelly
< Back to Terminator Measurement Help

|
Jason Kelly, as a graduate student in Drew Endy's lab, characterized transcriptional terminators BBa_B0011, BBa_B0014, BBa_B0015, BBa_B0021, BBa_B0024, and BBa_B0025 using his screening plasmid 1.0 pSB1A10. The following text describing his work is excerpted from Jason's PhD thesis, Tools and reference standards supporting the engineering and evolution of synthetic biological systems from MIT in 2008. |
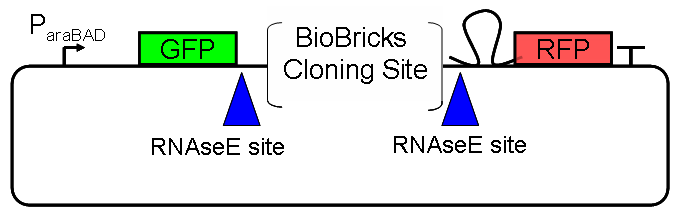
I used pSB1A10 (Figure 1) to measure the termination efficiency of a set of three BioBrick transcription terminators in the forward (BB_B0011, BBa_B0014, BBa_B0015) and reverse direction (BBa_B0021, BBa_B0024, and BBa_B0025). These terminators were chosen since they are used across many engineered biological systems built using BioBrick parts, in particular BBa_B0015 is one of the most widely used parts in the Registry (Randy Rettberg, personal communication). The termination efficiency was determined by measuring the per cell GFP and RFP fluorescence with a flow cytometer (Methods) after growing cells under a single induction level (0.003% arabinose). Terminator efficiencies were calculated by the following formula described previously Choe:
Termination efficiency = 1 - {[RFPterm/GFPterm]/[(RFPcontrol/GFPcontrol)mean]}
Where the subscript term refers to the measured GFP and RFP fluorescence when pSB1A10 contains a terminator and the subscript control refers to the measured fluorescence from pSB1A10 when no part is inserted in the insertion site (equivalent to 0% termination efficiency). The ratio of RFP to GFP fluorescence after background subtraction was averaged across all cells in the population for the control (denoted by the mean superscript). The ratio of RFP to GFP fluorescence after background subtraction was calculated for each cell in the population of cells containing pSB1A10 with a terminator inserted. The mean measured termination efficiency across all cells in the population for each terminator is provided in table 2-1. Since I measured the termination efficiency for each individual cell (Figure 2a), I also determined the distribution of termination efficiencies across cells in the population (Figure 2b).

In order to characterize the input / output relationship of a part or device using pSB1A10 I assume that the measured GFP and RFP fluorescence are directly related to the PoPS input and output of the part or device. However, in the case of transcription terminators this assumption is not fully justified. When a read-through event occurs (necessary for RFP to be expressed), a new hairpin is introduced to the 5’ end of the RFP mRNA transcript due to the terminator hairpin itself being part of the transcribed mRNA. It is known that 5’ hairpins are capable protecting transcripts from RNAseE cleavage Nicholson, and as a result it is possible that the RFP mRNA transcript is not being cleaved at the engineered RNAseE site within pSB1A10. Without this cleaving event the insulation between the particular part being measured and the downstream reporter (RFP) may be lost. This can have an unpredictable impact on the stability of the RFP mRNA transcript and will increase the error in terminator efficiency measurements. Future work is needed to better predict mRNA degradation rates from primary sequence or to design flanking sequences that might fix the degradation rates of mRNA at one predictable value Smolke.
References
<biblio>
- Khlebnikov pmid=12080425
- Smolke pmid=12402322
- Choe pmid=15767274
- Nicholson pmid=10371039
- Nojima pmid=17150951
</biblio>

