Help:Sequence Analysis Tools
- Registry Help Pages:
- TOC
- At-a-Glance
- FAQ
How we sequence parts
Part samples are sequenced using the VF2 and VR primer sites on their plasmid backbones. The VF2 and VR sites are located on BioBrick plasmids (just one of the reasons why you must send your parts in pSB1C3!). This ensures that the sequence reads will be able to read the BioBrick prefix and suffix as they move inward into the part sample.
- The VF2 and VR reads should be able to locate the BioBrick prefix and suffix, respectively (Ex. BB Prefix found at __).
- The quality of the reads begins to drop after several hundred bases. If the part is too long (generally >1kbp) the reads will either not reach into the middle of the part or their overlapping sections will have poor quality.
- Currently, we only do a single VF and VR read.
Sequence results are then uploaded and compared to their target sequence (part's documented sequence) through Registry software, and are automatically assigned several qualitative values.
Note: We are continuously looking to improve the Registry software and our sequence analysis. If you have any suggestions please send an email to hq AT igem DOT org.
Sequence analysis
To see if a part's sample has sequencing results, you can click the "Get This Part" link at the top of the page, and find if a part's sample/location has QC results. If so, click on the Sequencing link, right beside the part's Sequence result. This will take you to the Sequence Analysis page for your part.
![]()
We manually reevaluate "Inconsistent" sequences to see if the result was simply a software issue rather than the actual sequence being incorrect. Once a part has been re-evaluated by us or another user, a [U] is added its qualitative value. When possible we also leave comments in regards to the part's status.
Confirmed - The prefix and suffix is correct. The part's sample matches the part's documented sequence.
Long Part - The prefix and suffix is correct. The part's sample is visibly correct, however due to the limited length of the sequencing reads, the middle of the sample is not covered well or at all.
Partially Confirmed - The prefix and suffix is correct. An error may be present, but cannot be determined. (Quality of read may be poor; One read may show an error, while the other read cannot confirm it)
Questionable - The part's sample appears correct with one or two possible mutations. The mutations cannot be confirmed by both VF and VR reads.
Inconsistent - The part's sample does not match the part's documented sequence (as proved by both reads). The prefix and/or suffix may be incorrect.
Bad Sequencing - The sequencing reads are bad (poor quality).
No Target Part -
No Part Sequence - The sequence of the part has not been specified.
Note: We have begun to reevaluate all sequence reads
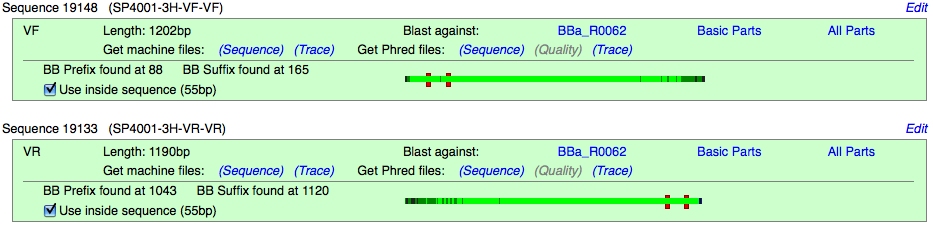
Current Sequence Analysis
The Current Sequence Analysis is the header section of the Sequence Analysis page. This section will let you:
- Click on the Target part: (BBa_R0062) to return to its main page
- See which source this sequence information is tied to (Linked to sample:)
- See the length: of the part in bp (length: 55bp)
- See the composition of the part; any subparts that may comprise the part
VF2 and VR Reads
The Registry sequences part samples using the VF2 and VR primer sites on the plasmid backbones. These reads are uploaded to the Registry and given an ID number (Ex. 19148 for VF). For each read you can:
- See the length of the sequence read
- See where the BioBrick Prefix and Suffix have been located, and if they have any errors
- See the size of the inside sequence, (length in between the prefix and suffix)
- Download sequence and trace files
- Sequence files are output as text
- Trace files can be viewed by 4Peaks(Mac) or DNA Tools Xplorer(Windows)
- Blast the read against the documented sequence of the target part (BBa_R0062), Basic Parts (non-composite parts), or All Parts on the Registry.
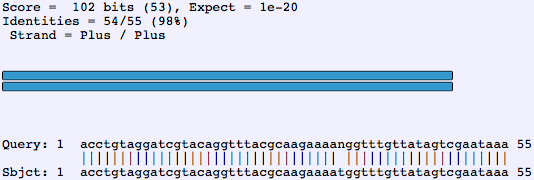
Blasting a Read
A blast against the target part can be looked over for inconsistencies between the subject (documented sequence) and the query (sequence of the read).
Vertical lines indicate a nucleotide match between the read and the target part's sequence. Inconsistencies are denoted by the lack of this vertical line between the two sequences, which indicates that:
- the bases disagree
- the sequencing was unable to determine a definite base in the query (the "n" in the above example)
- there is an additional or absent base(s) in the query
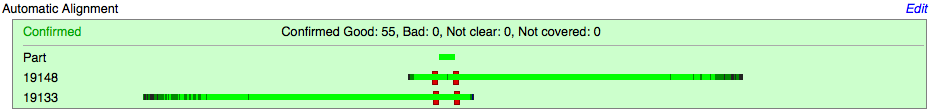
Automatic Alignment
The Automatic Alignment section:
- Displays a visual alignment of both reads to the target part.
- Sequence result decided by the Registry software or a user [U]: Confirmed , Inconsistent, etc.
- Any comments made by a user in regards to the part's sequence
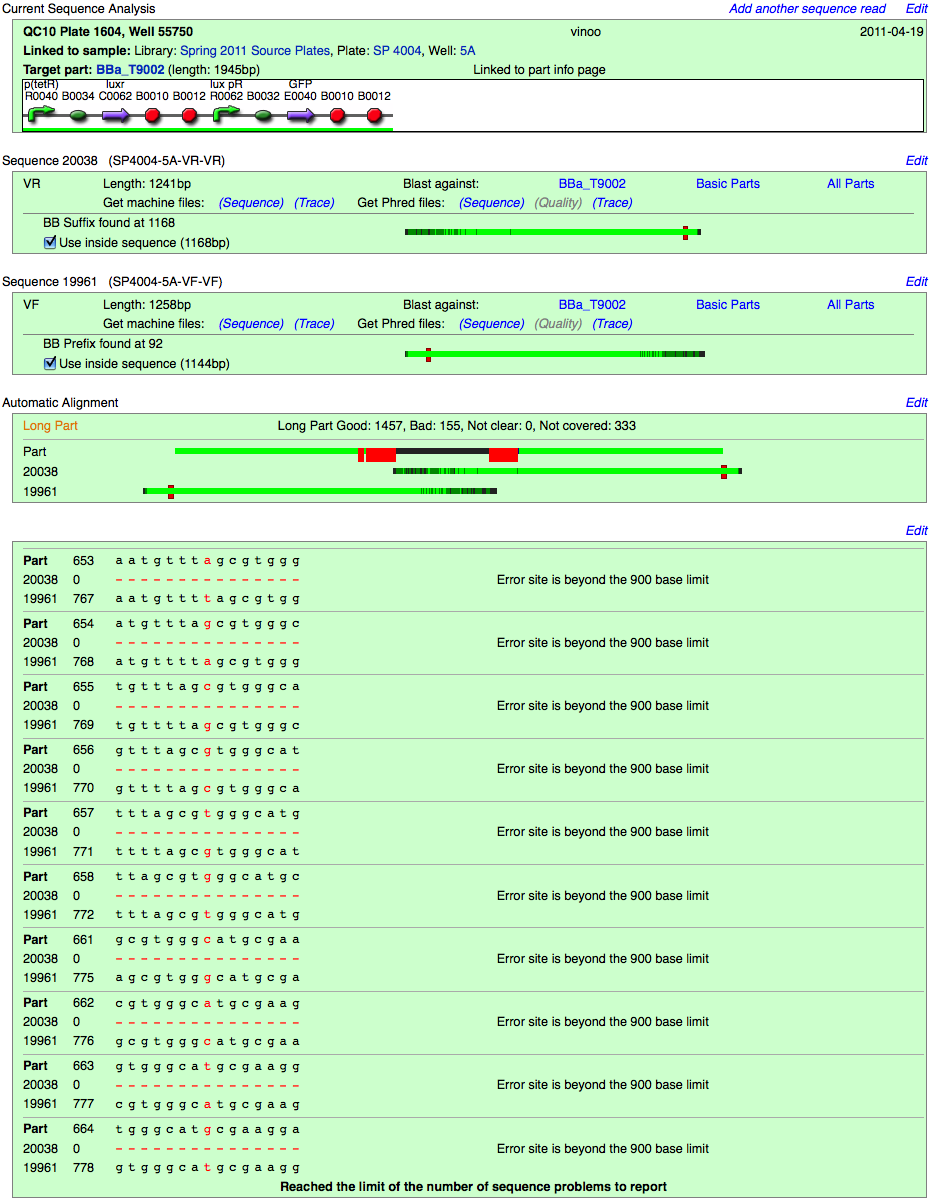
Sequence Problems
Below the automatic alignment, the Registry software will display Sequence Problems, specific nucleotide discrepancies between the reads and their target part.
The nucleotide in question is colored red, and its locations on the documented sequence of the part, and the two reads are noted.
Note: The Registry software has a limit of displaying 10 sequence problems.
Sequence Analysis Examples
Long Part
The length of the sequence reads are insufficient to cover the middle of the part. The ends are confirmed, but middle is not.
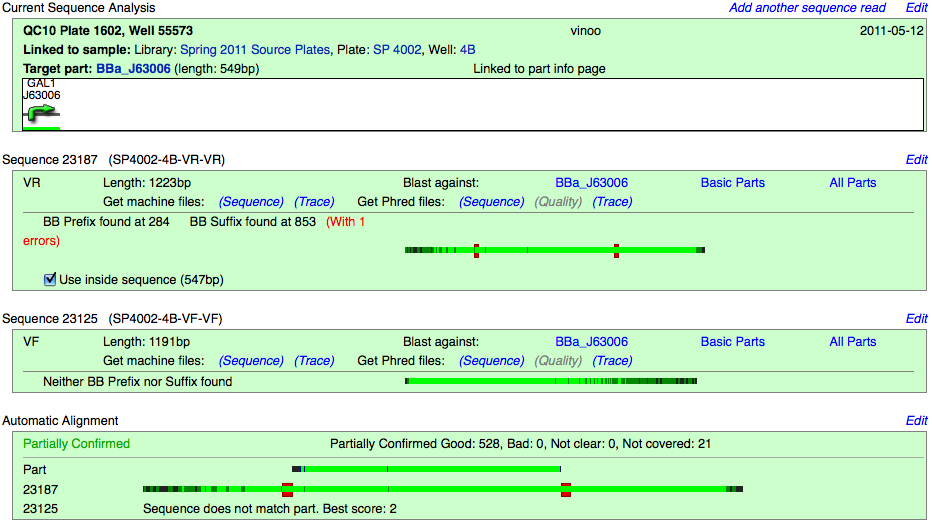
Partially Confirmed
The software is only able to partially confirm the sequence, in many cases this is likely due to one read being poor.
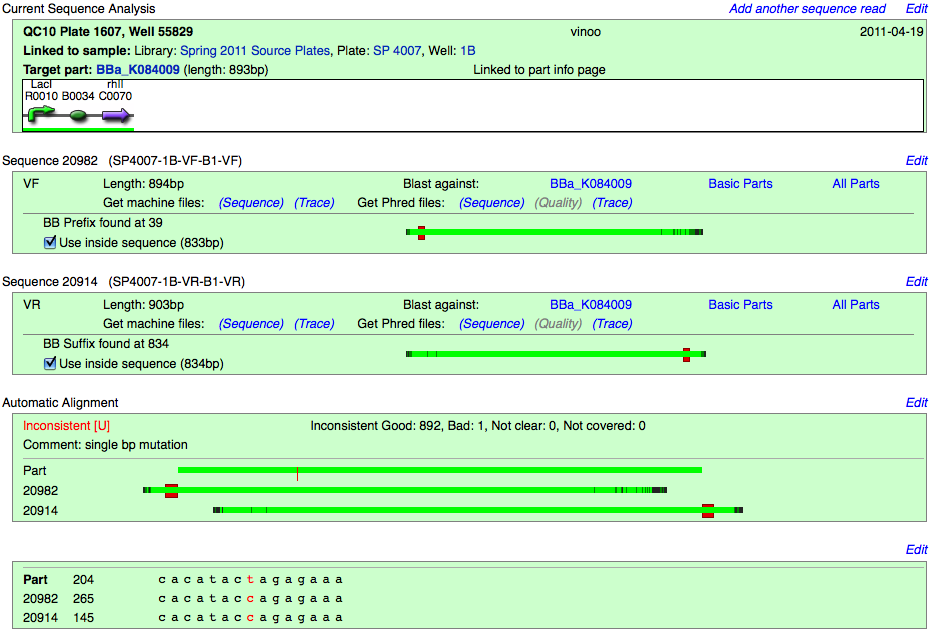
Inconsistent with mutation
The part matches mostly however a mutation(s) exists. Parts are still marked as Inconsistent, but comments will reflect the nature of their inconsistency.
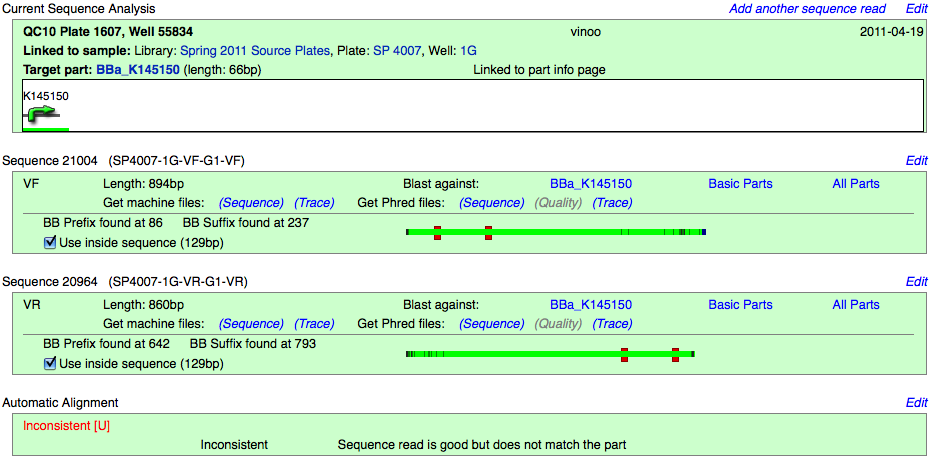
Inconsistent with no match
The part does not match at all.