Help:Sequence Analysis Instructions
Sequence Analysis Instructions
In preparation for iGEM 2008, a comprehensive quality control project was undertaken to ensure teams understood the quality of the parts before they began construction of their biological systems. DNA extracted from single colony culture was both digested and run on a gel, and sent to the Broad Institute for sequencing. The sequencing information received back from the Broad Institute was then uploaded into iGEM's software program and analyzed, comparing the sequences derived from our QC process to the sequences provided by the part designer. Additionally, each separate sequence was examined, and in some cases edited, by a lab technician to further ensure the sequences were correctly read by the software. By reading the following instructions, iGEM participants will be able to understand how the sequences were analyzed and pass their own judgment on the quality of the part.
- Go to the Registry of Biological Standard Parts webpage https://parts.igem.org/Main_Page
- Enter part ID# into Search option at top of the page
- Choose 'Physical DNA' option from heading
- Under 'Existing Sequence Analysis' heading, choose 'QC08 Plate ### Well #####'
Sequence Analysis Page
There are several important parts to the sequence analysis page and the following will explain the importance of each when analyzing DNA quality.
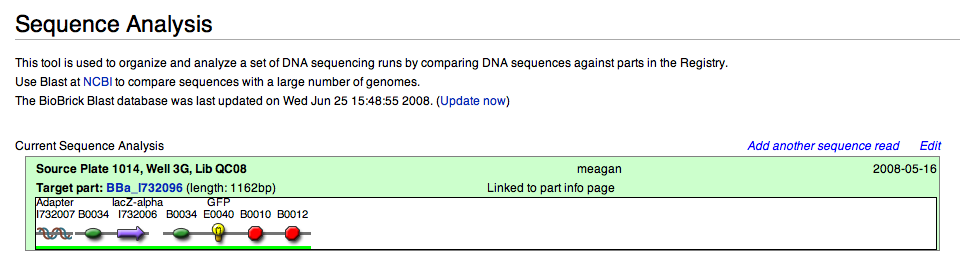
The Current Sequence Analysis box provides:
- source plate and well location in the Registry
- target part ID# with link to part page
- length (in bp) of part
- Subparts that comprise the part and links to information page
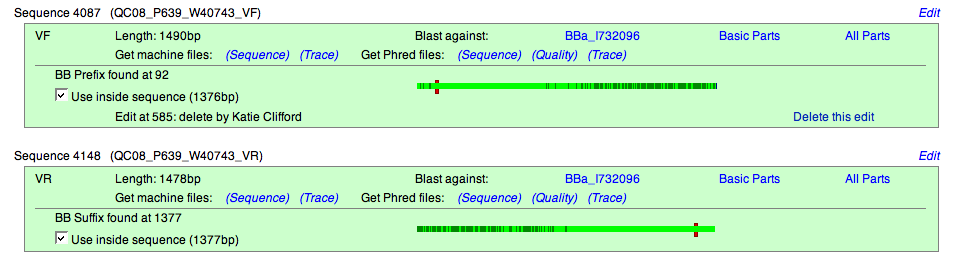
There are also two Sequence boxes - one for the forward read and one for the reverse read. Each of these boxes contain:
- length of the sequence read
- base where the BioBrick prefix or suffix was found
- sequence read in listed base pairs
- sequence read in trace file form
- Blast option - comparison of our sequence found during QC process to the sequence provided by the part designer
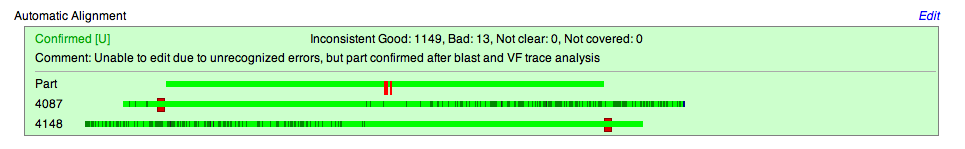
The Automatic Alignment box provides:
- Judgment passed on part quality
- a [U] icon, indicating editing
- post analysis comments
- comparison of part to both the forward and reverse sequence read
Analysis
- Open the blast results by choosing the highlighted part ID in either the forward or reverse 'Sequence' box
- Open the corresponding trace files by choosing 'Get Machine Files: (Trace)'
Note: You must have downloaded either 4Peaks(Mac) or DNA Tools Xplorer(Windows) programs to view trace files
- Choose 'View Chromatogram'
Once able to view both the blast and its corresponding chromatogram, the blast can be looked over for inconsistencies between the subject (documented sequence) and the query (sequence found during QC project).
Inconsistencies are denoted by the lack of a short vertical line between the two sequences. This indicates either:
- bases disagree
- sequencing was unable to determine a definite base in the query
- additional or absent base(s) in the query
When a discrepancy is found on the blast, that base location should be found on the corresponding trace in order to make sure the error found in the query is, in fact, shown on the trace.
The terms we used to classify the sequence reads are
- Confirmed- the sequence read matches the part sequence stored in the registry
- Partially confirmed- the sequence read obtained matches the part sequence stored in the registry with few exceptions
- Inconsistent- the sequence does not match the part sequence stored in the registry
- Questionable- undetermined if the part is confirmed
- Long part- the part was too long to obtain a full sequence read
- No target part- no sequence read was obtained
- No part sequence- no sequence was stored in the registry