Help:Ribosome Binding Sites/Standard Assembly
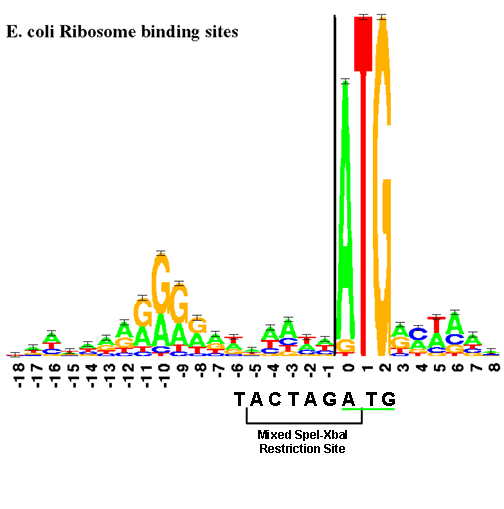
The RBS is located near the start codon. The "sequence logo" (similar to a consensus sequence), for E. coli RBSs is shown below. The size of each letter is proportional to the frequency of that base in the RBS part.
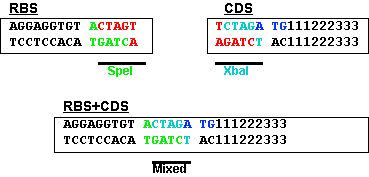
This figure shows the RBS is an A/G-rich region about 10 bases upstream of the start codon. There is little room for a BioBrick restriction enzyme site between the RBS and the start codon. BioBrick Standard Assembly 1 results in an 8-base region containing a 6-base mixed SpeI/XbaI restriction site surrounded by a base on each end (--T ACTAGA G--). Placing this sequence between an RBS and a start codon would lead to weak translation initiation since the spacing between RBS and start codon would be too large.
This problem is minimized by changing the rightmost base from a G to an T. This results in the sequence shown below and limits the impact of the BioBrick overhead to six bases.