Help:Ribosome Binding Sites/Mechanism
Contents
How do bacterial Ribosome Binding Sites work?
The bacterial ribosome binds to particular sequences on an mRNA, primarily the Ribosome Binding Site (RBS) and the start codon (AUG). The RBS base-pairs with an RNA molecule that forms part of the bacterial ribosome (the 16s rRNA), while the start codon base-pairs with the initiator tRNA which is bound to the ribosome. In addition to the sequences of the RBS and the start codon being important, these two sequences need to be positioned approximately 6-7 nucleotides apart so they can both make contact with the appropriate parts of the ribosome complex. In the next section, we talk more about the specific interaction between the RBS and the ribosome.
The Shine-Dalgarno Sequence
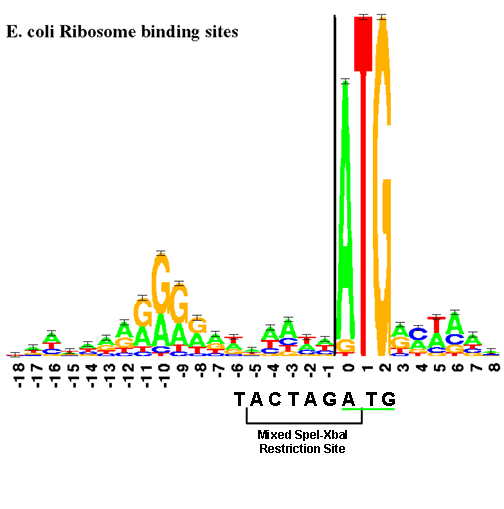
The end of the 16s rRNA that is free to bind with the mRNA includes the sequence 5′–ACCUCC–3′. The complementary sequence, 5′–GGAGGU–3′, named the Shine-Dalgarno sequence, can be found in whole or in part in many bacterial mRNA. Very roughly speaking, ribosome binding sites with purine-rich sequences (A's and G's close to the Shine-Dalgarno sequence will lead to high rates of translation initiation whereas sequences that are very different from the Shine-Dalgarno sequence will lead to low or negligible translation rates. The sequence is named after the researchers who discovered the high frequency of the sequence upstream of bacterial coding sequences Shine. You can see how common the sequence is by looking at the RBS sequence logo on the right (where the height of a letter indicates the frequency of the letter at that location).
How do eukaryotic Ribosome Binding Sites work?
Initiation of protein synthesis in eukaryotes differs from this model. The 5' end of the mRNA has a modified chemical structure ("cap") recognized by the ribosome, which then binds the mRNA and moves along it ("scans") until it finds the first AUG codon. A characteristic pattern of bases (called a "Kozak sequence") is sometimes found around that codon and assists in positioning the mRNA correctly in a manner reminiscent of the Shine-Dalgarno sequence, but not involving base pairing with the ribosomal RNA.
References
<biblio>
- Shine pmid=803646
- Shultzaberger pmid=11601857
- Vellanoweth pmid=1375309
</biblio>