Help:DNA/Glossary
Contents
BioBrick cloning site
| To adhere to the BioBrick standard for physical composition of genetic parts, plasmid backbones must include a BioBrick cloning site. The cloning site includes a BioBrick prefix sequence composed of an EcoRI, NotI, and XbaI restriction enzyme recognition site and a BioBrick suffix sequence composed of a SpeI, NotI, and PstI restriction enzyme recognition site. | |
| In most BioBrick plasmid backbones, including those constructed from the BioBrick base vector, the BioBrick prefix and suffix sequences are not adjacent to one another. Instead, the BioBrick prefix and suffix flank what is a called a default plasmid insert or default insert for short. There are several different default inserts available in the Registry. See default plasmid inserts for more information. |
DNA aptamers
Nucleic acid aptamers are single- or double-stranded DNA or RNA sequences than can bind specifically to molecular targets including but not limited to small molecules, proteins, and organims. Aptamers offer a popular platform for generating specific and high affinity ligands, because large aptamer populations can be randomly generated via chemical synthesis, screened for binding through in vitro selection, and amplified/diversified by polymerase chain reaction Ellington,Gold. Nanomolar dissociation constants can readily be obtained for molecular targets. Nucleic acid aptamers are best suited for molecules such as basic proteins. Nonpolar molecular targets offer a far more difficult target for aptamers, because the nucleic acid phosphate backbone is negatively charged.
One of the most popular methods for obtaining specific and high affinity aptamers to a particular molecular target is systematic evolution of ligands by exponential enrichment (SELEX) Gold. SELEX involves iterative cycles of aptamer selection and amplification, either with or without intentional random point mutagenesis. David Liu's group has also published on another technique for identifying specific and high affinity aptamers through nonhomologous random recombination (NRR) that may offer even better results than SELEX Bittker.
Andy Ellington's lab at the University of Texas at Austin maintains a database of published nucleic acid aptamer sequences at http://aptamer.icmb.utexas.edu/.
DNA origami
DNA offers an attractive material for constructing nanostructures. In particular, the high specificity of Watson-Crick base pairing allows a diverse set of binding interactions to occur in parallel in a reasonably predictable fashion Watson. Several groups have leveraged this feature of DNA to construct both two-dimensional sheets and three-dimensional structures Seeman, Shih. In 2006, Rothemund built upon previous work to demonstrate a general method for fabricating DNA into any two dimensional shape Rothemund. His algorithm generates short synthetic DNA staples that direct the folding of a long single-stranded DNA molecule (cloning vector M13 mp18) into arbitrary shapes.
Plasmid replication origin
The genetic element responsible for the replication of plasmids during cell growth and division is called a replication origin (also "origin of replication" or simply "origin"). There are several different replication origins and they differ in their plasmid copy number per cell (how many molecules of the plasmid are maintained in the cell), mechanism of copy number control, cell-to-cell copy number variation, and even the degree of coiling of the physical DNA. Thus, BioBrick® parts, devices and systems can operate very differently from one plasmid backbone to another.
| Several replication origins have been designed in the Registry. Although most of them are not available as individual BioBrick parts, some can be obtained by PCR of existing plasmid backbones in the Registry using appropriate primers. In fact, when making new BioBrick vectors using the BioBrick base vector, Reshma simply assembled a PCR-amplified linear DNA encoding the replication origins with the antibiotic resistance markers. (The PCR primers included the BioBrick prefix and suffix so that the linear DNA encoding the origin was ready for digestion with EcoRI and SpeI. |
Primer binding sites
| Primer binding sites are DNA sequences specifically designed for oligo annealing. For example, most BioBrick plasmid backbones include binding sites for the VF2 and VR primers. | |
Recombination sites
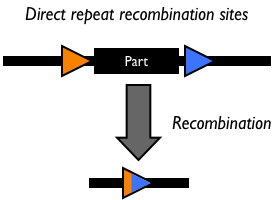
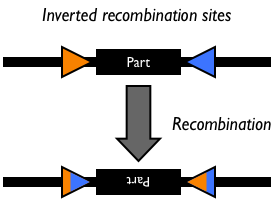
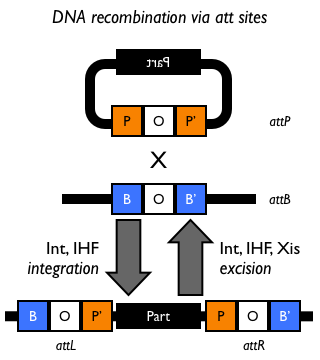
Site-specific DNA recombination requires both a recombinase protein and a pair of repeated DNA sites at which recombination takes place. Depending on the number and orientation of the DNA sites, there can be either an inversion, deletion or insertion of DNA (Figure 1). While some DNA recombination systems, such as Cre/lox only require the recombinase and the two DNA sites for recombination to occur, others either require or are modulated by additional accessory factors.
Figure 1: Schematic of different types of recombination events.
For more information on particular DNA recombinases and their target sites, see DNA recombination sites.
Transposons
Transposons are sequences of DNA that can move around to different positions within the genome of a single cell. Together with transposases, transposons make up transposomes. Transposomes are frequently used to construct gene knockouts in living cells which can be helpful for identifying essential genes.