Chassis/Cell-Free Systems/Homemade E.coli S30/Preparation Protocol
Contents
Homemade E. coli S30
| Cell-Free Systems | Chassis description | Preparation protocol | Preliminary testing |
Preparation Protocol
Day 1
Equipment
- 37°C shaking incubator
- 1L conical flasks x 3
- Pipette fillers + pipettes (5ml, 10ml and 25ml)
- Spectrometer + cuvettes
- Weighing scale
- Centrifuge + 150ml centrifuge tubes
- Pipettes + pipette tips (20µl, 200µl and 1000µl)
Reagent
- 2xYT medium
- IPTG
- Buffer A
- 10mM Tris-acetate (pH 8.2)
- 14mM Mg-acetate
- 60mM K-lutamate
- 1mM dithiothreitol (DTT)
- 0.05% (v/v) 2-mercaptoethanol (2-ME)
Procedure
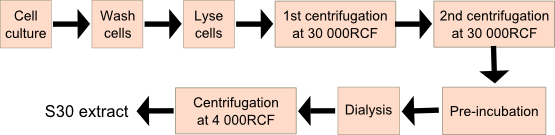
Growing the cells
- Grow E. coli strain BL21 (DE3) cells at 37°C in 3L of 2xYT medium till O.D.600 = 0.6.Ensure vigorous agitation and aeration.
- Add 1mM IPTG to cell culture to express T7 RNA polymerase.
- Harvest cells when O.D.600 = 4.5. At this point, cells are at mid-log phase.
- Wash cells three times by suspending them in 20ml of buffer A per gram of wet cells.
- Centrifuge and weigh the wet cell pellets before storing them at -80°C.
(Note: The cells may take more than 1 day to grow to O.D.600 = 4.5.)
Day 2
Equipment
- Pipette filler + pipettes (5ml, 10ml and 25ml)
- Weighing scale
- French press + French press cell
- Centrifuge + 50ml centrifuge tubes
- Pipette + pipette tips (20µl, 200µl, 1000µl)
- 37°C shaking incubator
- Dialysis membrane with molecular weight cut-off of 10,000
- Magnetic stirrer
- 4°C cold room
Reagent
- Buffer B
- 10mM Tris-acetate (pH 8.2)
- 14mM Mg-acetate
- 60mM K-glutamate
- 1mM DTT
- Pre-incubation solution
- 293.3mM Tris-acetate (pH 8.2)
- 2mM Mg-acetate
- 10.4mM ATP
- 4.4mM DTT
- 0.04mM amino acids
- 16.9mM phosphoenolpyruvate
- 0.77U/ml pyruvate kinase
(Note: For the ATP regenerating system in the pre-incubation solution, phosphoenolpyruvate and pyruvate kinase are used instead of creatine phosphate and creatine kinase. This is due to cost considerations.)
Procedure
Lysing the cells
- Suspend thawed cells in 12.7ml of buffer B per 10g of wet cells.
- Disrupt cells in a French press cell at a constant pressure of 20,000psi.This is about 140,000kPa.
Retaining the cell extract
- Centrifuge the crude lysate at 30,000RCF for 30min at 4°C.
- Carefully remove the top layer of the supernatant (lipid layer) and the pellet and centrifuge again.
- Shake the final supernatant at 100rpm.
- Gradually add 3ml of the pre-incubation solution to 10ml of the supernatant.
- Pre-incubate the supernatant with gentle shaking at 37°C for 80min. This degrades endogenous genetic content (DNA and mRNA).
- Dialyze the pre-incubated sample for 45min each at 4°C against 50 volumes of buffer B using a membrane with molecular weight cut-off of 10,000. Repeat the dialysis step three times.
- Centrifuge the retained extract at 4000RCF for 10min at 4°C to obtain the supernatant.
- Divide resulting S30 extract into small aliquots and store at -80°C.
(Note: Protease inhibitors are added to pre-incubation solution to prevent degradation of proteins required for gene expression.)
Notes
- Total time required: ~ 3 days.
- The protocol is based on Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system by Kim DM et al.
- Modifications to protocol:
- The original protocol uses creatine phosphate and creatine kinase instead of phosphoenolpyruvate and pyruvate kinase.
- The original protocol does not use protease inhibitors in the pre-incubation solution.
The cell-free protein synthesis reactions will be carried out in a 1.5 mL microtube placed in a water bath set at 37 °C.
Reaction mixture
- 57 mM Hepes–KOH (pH 8.2)
- 1.2 mM ATP
- 0.85 mM each of CTP, GTP and UTP
- 2 mM DTT
- 0.17 mg/mL E. coli total tRNA mixture
- 90 mM potassium glutamate
- 80 mM ammonium acetate
- 12 mM magnesium acetate
- 1.5 mM each of 20 amino acids
- 2% PEG (8000)
- 110 mM phosphoenolpyruvate
- 15 U/ml pyruvate kinase
- 6.7 μg/mL DNA
- 27% (v/v) of cell extract
References
<biblio>
- 1 pmid=16797767
</biblio>