Chassis/Cell-Free Systems/Homemade E.coli S30/Preliminary Testing
Contents
Homemade E. coli S30
| Cell-Free Systems | Chassis description | Preparation protocol | Preliminary testing |
Preliminary Testing
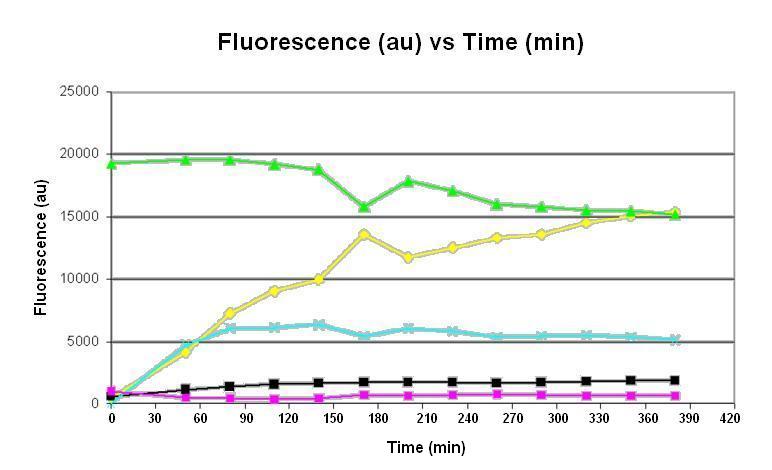
Graph 1. Graph of fluorescence (au) against time (minutes). The pink line represents the negative control consisting of homemade E. coli S30 extract and homemade premix with no DNA; the light blue line represents the reaction using homemade E. coli S30 extract and homemade premix with DNA; the yellow line represents the reaction using commercial E. coli S30 extract and commercial premix with DNA; the black line represents the reaction using homemade E. coli S30 extract and commercial premix with DNA; the green line represents the positive control consisting of GFP protein in solution. The inducible gene expression device BBa_T9002has been used.
Experimental Protocol
Aims
- To perform preliminary testing on the homemade E. coli S30 chassis.
- To compare the homemade E. coli S30 chassis with the commercial E. coli S30 chassis.
Equipment
- Fluorometer + Connected PC Turn on before beginning
- 96 well plate x1 + Plate lid
- 1.5ml eppendorf tube x7
- eppendorf rack
- Gilson p20,p200,p1000
- Stop watch
Reagents
- Homemade E.coli S30 extract
- Commercial E.coli S30 extract. Including:
- 175µl Amino Acid Mixture Minus Cysteine, 1mM
- 175µl Amino Acid Mixture Minus Methionine, 1mM
- 175µl Amino Acid Mixture Minus Leucine, 1mM
- 450µl S30 Extract, Circular (3 × 150µl)
- 750µl S30 Premix Without Amino Acids
- MiiA water x1ml
- GFP solution (For this initial experiment does not need to be purified GFP, we just want to know we have the right filter and that our settings are adjusted to measuring GFP)
Preparation
- First collect all equipment and reagents and ensure that the fluorometer and that the PC connected has a data collection protocol installed.
- Commercial E.coli Cell Extract: First prepare a complete amino acid mixture for both extract solutions: Add the 7.5μl volume of two amino acid minus mixtures into an labeled eppendorf to give a volume of 15μl. Each amino acid minus mixture is missing one type of amino acid, and so by combining two solutions we are complementing each solution for the missing amino acid. Take a eppendorf tube and add 5µl of the E.coli complete amino acid mixture. Then add add 20µl of S30 Premix Without Amino Acid. Then add 15µl of S30 Extract Circular. Finally add nuclease-Free Water to bring final volume (inc.DNA vol) to 60µl, the volume of DNA added will be determined in experiment 1 and the volume of the nuclease free water adjusted accordingly. Place the eppendorf tube in a rack on the bench
- Homemade S30 Cell Extract: Reaction mixture consists of S30 cell extract (16.2µl), reaction buffer (30µl),puruvate kinase (3.1µl), rNTPs (1µl), DNA (4µl) and ddH2O (5.7µl), making up a total volume of 60µl. Place these tubes in a rack on bench.
Plate loading
- Begin by loading the in vitro expression system into the correct wells. Before loading in the samples vortex the tubes for a few seconds to mix the solution.
- Place the lid on the 96well plate and tape up the edges of the lid. This should be put into the incubator at 37oC for 10 minutes to allow temperature to equilibrate
- Remove from 37oC incubator and spin-down in centrifuge in plate centrifuge at 2000rpm for a few seconds. Spin down is the process of bringing down any solution on lid or side of well into the base of the well. Alternatively can tap the top of the lid to bring down any solution to bottom of the well.
- Remove lid off th e 96well plate and place in the fluorometer. Once the data collection is set up then initiate the measurements.
- This measurement will give a background fluorescence measurement and can be used as our time zero data.
- Then to begin the reaction add required volume of purified DNA sample to give 2µg to the appropriate wells. Be careful not to add to wells that DO NOT NEED DNA.
- Place lid back on and place back in the incubator at 37oC.
- After 30 minutes of incubation measure the fluorescence by repeating procedure 3-4 above. This initial measurement of 30 minutes is to find out how fast GFP is being produced. After this initial measurement, the intervals should be reassessed and adjusted accordingly.
- Before each measurement be careful to remember to either spin down or tap down the solution and to remove the lid before placing in the fluorometer.
- Continue taking measurements for 6h.
- Plot a graph of fluorescence against time for the various samples.
