Part:BBa_K771308
FabI with Membrane Anchor 5(without VVD )
BBa_K771308:FabI is linked to C terminal of BBa K771008 by Flexible Linker 3(FL3).
Fatty acid Biosynthetic Pathway
TesA involves in the biosynthesis of fatty acids in E.coli. The biosynthesis pathway is shown as below:
The E. coli fatty acid synthesis is initiated when holo-ACP, NADPH and NADH, acetyl-CoA and malonyl-CoA undergo condensation and subsequent reduction to form butyryl-ACP. These reactions are catalyzed by the malonyl-CoA:ACP transacylase FabD, the ketosynthase FabH, the NADPH-dependent ketoreductase FabG, either the dual-function dehydratase/isomerase FabA or the monofunctional dehydratase FabZ, and the NADH-dependent enoyl reductase FabI. Then the butyryl-ACP is extended via 5-7 rounds of analogous reactions to produce a C14 to C18-ACP either fully saturated or monounsaturated. These extension cycles are catalyzed by either the ketosynthase FabB or FabF in collaboration with FabD, FabG, FabA or FabZ, and FabI. Finally, the full-length fatty acid is released from the corresponding fatty acyl-ACP via hydrolysis by C16-specific thioesterase,TesA.
Design of Experiment
Mechanism beneath our idea suggests the Membrane Magic project enjoys two fundamental privileges: the Priority to Exportation and the Refinement of Interaction.
The Priority to Exportation
The fatty acid synthesis is terminated by the hydrolysis of fatty acyl-ACP and the release of free fatty acids into cytoplasm. We suppose TesA anchored on membrane could effectively increase the concentration of fatty acids near membrane, which in turn, facilitates the transmembrane transportation of fatty acids. Higher level of fatty acids in the culture medium make it easier to obtain and purify product and more suitable for Industrialized production. On the other hand, TesA removes fatty acyl-ACP from the reaction and thus, according to Le Châtlier principle, shifts the chemical equilibrium to accelerate and to accumulate more fatty acids.
To identify and to evaluate the priority of products to exportation, controlled experiments were designed and conducted. Wild type E.coli and E.coli expressing free-diffuse TesA were used as control groups. TesA was fused with transmembrane protein No.1 as stated in the Construction and the fusion protein was expressed in experimental group.
The Refinement of Interaction
Enzymes fused with membrane anchors will be directed to the membrane as expected right after their translation. Thus, the distribution of enzymes is restricted to the 2D membrane rather than diffusing randomly throughout the cytoplasm. Due to spatial restriction, we expect the receptor-ligand interaction that we employ to form enzymes cluster occurs at a higher frequency comparing to scaffolds in other forms. Moreover, the interaction could be stabilized by phospholipid around transmembrane domain. We believe a tandem of enzymes involved in sequential reactions could be established on the membrane swiftly and orderly.
To testify and to assess the refinement of interaction, controlled experiments were designed and conducted. Four enzymes are selected based on previous study. We express thioesterase, TesA and full complement of reductive enzymes, FabG, FabZ and FabI because moderate overexpression of TesA in gave rise to elevated fatty acid productivity and reductive enzymes could lead to 50% increase in fatty acid turnover. Wild type E.coli and E.coli expressing those four free-diffuse enzymes were used as control groups. TesA was fused with transmembrane protein No.1 as stated in the Construction and FabG, FabZ and FabI with No.2 to No.4 respectively, align with corresponding reactions occurring in sequence. Four fusion proteins were expressed together in experimental group.
Result and Discussion
Overview
Excitingly, the fatty acid biosynthesis was accelerated sharply by recruiting membrane accelerator. The introduce of TesA alone results in 50% increase in fatty acids yield. By gathering FabG, FabI, FabZ and TesA on the membrane, the production of fatty acid was enhanced significantly by 9 fold compared with free enzymes in cytoplasm. The statistical data result strongly supports the hypothesis above that Membrane Magic make it much easier for exportation and interaction.
To demonstrate the priority to exportation and the refinement of interaction, two sets of controlled experiments have been set as following:
WT stands for E.coli transformed with corresponding plasmid(s) without exogenous gene.
F- protein here indicates enzyme diffusing randomly throughout the cytoplasm without modification.
M- protein means enzyme fused with engineered transmembrane domine and localized to E.coli membrane where enzymes aggregate and cooperate.
The solo introduce of TesA with Membrane Anchor 2 (without MS2)
As Fig.4 A indicates, 20 hours after induction, E.coli with membrane anchored TesA experienced a 50% increase in C-16, C-18 fatty acids content and total fatty acids (Fig.4 B) in supernatant, compared with E.coli with free TesA.Fatty acids yielded from supernatant went up to 0.71mg/(L·OD). The result support that membrane anchored TesA could efficiently transfer fatty acyl-ACP into desirable fatty acids right beneath inner membrane, thus making it much easier for the final products to diffuse into periplasmic space.
Fig.5, on the other hand, shows fatty acids content in sedimentation also went up by 40%, up to 5.02mg/(L·OD). The result is still within our expectation since fatty acyl-ACP has been removed from the reaction system to form fatty acids. As a result, the chemical equilibrium shifts and more fatty acids would be accumulated.
The cooperation of enzymes with membrane anchors
TesA with Membrane Anchor 2 (without MS2), FabG with Membrane Anchor 3, FabZ with Membrane Anchor 4, FabI with Membrane Anchor 4(without VVD) could significantly enhance the production of fatty acid by 9 fold compared with enzymes without membrane anchor.
To optimize the productivity of the system we established, we tended to combine these two privileges together, gathering TesA, FabG, FabI and FabZ through receptor-ligand interaction. Notable increase in both diversity and amount of fatty acids were detected.
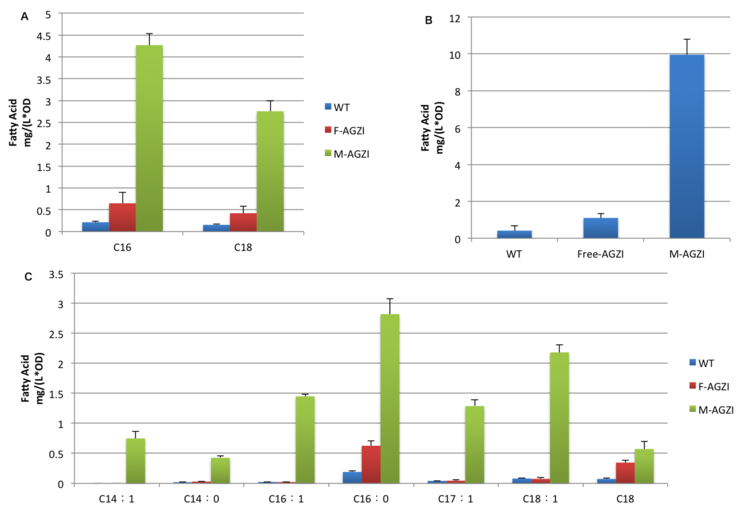
Cluster of FabG, FabI and FabZ provides C16- and C18 specific TesA with sufficient amount of fatty acyl-ACP to hydrolyze and release. Therefore, we witnessed a tremendous growth in the turnover of fatty acids with C16 and C18 skeleton in the supernatant.(Fig.6 A)
Moreover, fatty acids with C14 skeleton was first detected in E.coli expressing membrane anchored enzymes compared with ones with free enzymes. It is probably because the productivity of cluster of FabG, FabI and FabZ overloads itself when carbon chain growth slows down as it elongates. Monounsaturated fatty acids also emerge in considerable amount for the first time since TesA is located so closely to the cluster of FabG, FabI and FabZ that it catches intermediate with C16 and C18 skeleton even before they are reduced.(Fig.6 C)
Augment in both diversity and amount of fatty acids led to 24 fold increase in total yield compared with wild type and 9 fold with ones expressing free enzymes. (Fig.6 B) The exciting result convincingly proves that membrane enhances receptor-ligand interaction and cluster of enzymes makes it faster to a surprising extent. Products acclumulates near inner membrane and traval a shorter distance to diffuse through membrane.
The fatty acids increase in sediment is relevantly moderate. The reason might be that membrane anchored TesA exports large quantity of fatty acids so the amount remaining in intracellular is limited.C16 and C18 fatty acids in sediment experienced similar trend as that in supernatant and the reason may be the same. It is notable that C19 fatty acids only exist in inside the cell and experience unique changing pattern in which membrane anchored enzymes together produce even less than free ones. We suppose it is caused by the overuse of precursors which inhibits the elongation cycle. Large molecular weight of C19 fatty acids prevent it from exportation and finally they are trapped in the cell.
Protocols
- Transfer a fresh transformed colony to 5ml LB liquid medium and shake at 37℃ overnight(12h).
- Add seed broth to 50ml LB medium(volume ratio is 3% and loaded liquid is 20%) and shake until the OD600 reaches 0.6 or so. Add inducer of appropriate concentration to induce gene expression.
- Continue shake culture for another 20h. Contrifuge at 8,000 for 5min. Collect supernatant and bacteria sediment respectively (prepare 3 parallel samples, each 10ml).
- Extract the supernatant by chloroform-methanol(2:1) solution of the same volume (shake for 5min) place until stratification. Dry the lower organic layer at 50℃ for about 24h. Suspend the bacteria by 1ml ddH2O. Add 20ml chloroform-methanol solution and extract for another 2 times.(Attention: weigh the empty flask before drying).
- Weigh the crude extract and record the results.
- Dissolve the crude extract with 1ml methanol and add 2ml tetrafluoroboron. Methyl esterificate at 60℃ for 30min. After the reagent cools down, add 2ml N-hexane and extract for 2 times. Collect the upper layer and dry overnight at 50℃ .
- Add the crude bacteria extract to 1ml chloroform-methanol(2:1) solution and shake by vortex to extract the total lipid of the cells. Add 2ml methanol-water(4:1) as saponification reagent. Saponificate in 60℃ water bath for 1h. Then esterificate as the 6th step.
- Dissolve the obtained fatty acidmethyl ester(FAME)mixture with N-hexane. Take 1ml sample for test.
- GC-MS condition:
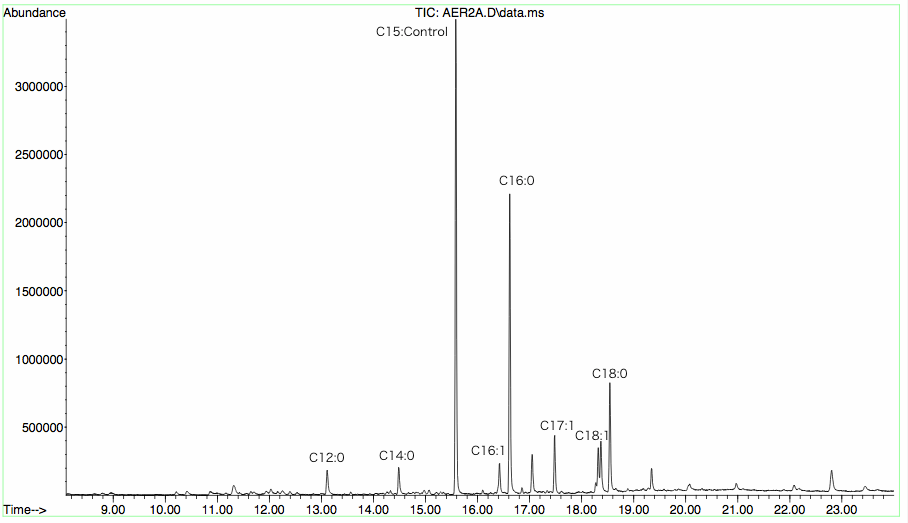
Sample injector temperature: 250℃; Detector temperature: 280℃; Temperature programming: begin at 100℃ and last 2min. Warm to 250℃ at the rate of 10℃/min and last 5min. Use C15 FAME as the interior label for quantification. (Quantity of C15 FAME depends on the sample concentration)
GC-MS
The solo introduce of TesA with Membrane Anchor 2 (without MS2)
Supernatant
Sediment
<br\><br\>
<br\><br\>
The cooperation of enzymes with membrane anchors
Supernatant
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 883
Illegal AgeI site found at 1237 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 504
| n/a | FabI with Membrane Anchor 5(without VVD ) |